| Journal of Food Bioactives, ISSN 2637-8752 print, 2637-8779 online |
| Journal website www.isnff-jfb.com |
Original Research
Volume 22, June 2023, pages 43-52
Protective effect of Melanogrammus aeglefinus skin oligopeptide in ultraviolet B-irradiated human keratinocytes
Ziyan Wanga, b, Lisha Dongb, Jiaojiao Hana, b, Jun Zhoua, b, Chenyang Lua, b, Ye Lia, b, Tinghong Minga, b, Zhen Zhanga, b, Rixin Wangb, *, Xiurong Sua, b, *
aState Key Laboratory for Quality and Safety of Argo-products, Ningbo University, Ningbo 315211, China
bSchool of Marine Science, Ningbo University, Ningbo 315832, China
*Corresponding author: Rixin Wang and Xiurong Su, School of Marine Science, Ningbo University, 169 Qixing South Road, Ningbo 315832, China. E-mail: wrx_zjou@163.comand suxiurong_public@163.com
DOI: 10.31665/JFB.2023.18347
Received: June 2, 2023
Revised received & accepted: June 17, 2023
| Abstract | ▴Top |
Ultraviolet B (UVB)-induced cell death causes skin photoaging. In this study, we investigated the protective effect of Melanogrammus aeglefinus skin oligopeptide (MSOP) in UVB-irradiated human keratinocytes. The method of preparing MSOP was optimized, and three peptides with high abundance, VADML (Val-Ala-Asp-Met-Leu), IARF (Ile-Ala-Arg-Phe) and SSPSF (Ser-Ser-Pro-Ser-Phe), were identified. Discovery Studio predicted that these peptides interacted with Keap1 and contributed to antioxidant activity. Therefore, a UVB-induced cell model was used to explore the beneficial effects of MSOP in vitro. The activities of superoxide dismutase and glutathione peroxidase were increased in the MSOP-treated groups, while the malondialdehyde content was decreased. In addition, 23 differentially expressed proteins were identified through quantitative proteomics analysis; among them, the upregulation of Nrf2 and downregulation of Keap1, which are involved in the Keap1/Nrf2/ARE signaling pathway, contributed to the antioxidant process. Based on this study, MSOP might be an alternative agent for protecting the skin against UVB exposure.
Keywords: Melanogrammus Aeglefinus skin oligopeptide; Ultraviolet B; Photic damage; Antioxidation; Quantitative proteome
| 1. Introduction | ▴Top |
Exposure to excessive sunlight is an important etiologic factor in the development of acute inflammation and is thus consequently linked to the progression of skin cancer (Hooda et al., 2023). Ultraviolet B (UVB) is the most damaging component of solar radiation that reaches the earth and is a major risk factor for the development of acute inflammation as well as nonmelanoma skin cancer in the epidermis (Lim et al., 2022). When the skin is exposed to UV light, a photooxidative reaction is initiated that impairs the antioxidant status and increases the cellular level of reactive oxygen species (ROS) (Kumar et al., 2022; Masaki et al., 2009). This process, which is commonly known as photoaging, impairs the self-protection property of the skin and consequently damages the cutaneous tissues (Berry et al., 2022).
Skin aging is a complex phenomenon that results from interactions between intrinsic and extrinsic factors. Intrinsic or chronological aging is an inevitable, genetically programmed process related to a natural decline in the normal physiological process whereas extrinsic aging is caused by environmental factors (Löwenau et al., 2017; Uitto, 1997). Both intrinsic and extrinsic factors are superimposed in sun-exposed areas of skin, resulting in marked histological changes in the extracellular matrix (ECM); clinically, the changes are observed as, photoaged skin with loss of rigidity and elasticity (Freitas-Rodríguez et al., 2017; Oh et al., 2020). Antioxidants have the capability to quench ROS and have been reported to repair skin damage, loss of elasticity, wrinkling and premature aging when supplemented in the diet (Fernandes et al., 2023; Liu, et al., 2023). Thus, it is suggested that regular consumption of antioxidant-containing natural ingredients might be a useful strategy for protection against UV-mediated cutaneous damage.
Cod is among the most important commercial food species used worldwide, and a large amount of byproduct is generated during processing. Skin is a good source of collagen, which accounts for 8% of the byproduct (Bruno Siewe et al., 2021; Chen et al., 2020). It has been reported that cod skin oligopeptide induces apoptosis and leads to inhibition of human gastric carcinoma cell growth (Wu et al., 2020), while a calcium-chelating peptide isolated from Pacific cod skin gelatin showed the ability to improve calcium absorption (Wu et al., 2017). In addition, active peptides derived from pacific cod skin demonstrated antioxidant activity and angiotensin-I converting enzyme inhibitory ability (Ngo et al., 2011). Moreover, further research that clarified the underlying mechanism was mediated by regulation of the MAPK and transforming growth factor-β/Smad signaling pathways (Chen and Hou, 2016; Chen et al., 2016). However, there is little information about the protective effect of Melanogrammus aeglefinus skin oligopeptide (MSOP) in UVB-irradiated human keratinocytes, and the underlying mechanism remains unclear.
In this study, the method of preparing MSOP was optimized, the composition and abundance of peptides in MSOP was identified by MALDI-TOF/TOF, the function of peptides was predicted by Discovery Studio 2019, the protective effect of MSOP was explored in UVB-irradiated human keratinocytes, and the underlying mechanism was investigated by proteomics. We proposed that MSOP has potential applications in UVB-induced photoaging.
| 2. Materials and methods | ▴Top |
2.1. Optimizing the method used to prepare MSOP
Pepsin (Shanghai Yuanye Bioengineering Institute, Shanghai, China) was selected to hydrolyze Melanogrammus aeglefinus skin collagen (Shanghai Nitta Gelatin Co., Ltd, Shanghai, China) according to previous studies (Song et al., 2022). Single-factor experiments were performed to establish the range of independent variables [temperature, enzyme/substrate (E/S) ratio and time]. Then Box-Behnken response surface design was formulated to evaluate and optimize the effects of these three independent variables and their interactions on the degree of hydrolysis (DH). The DH was measured by trichloroacetic acid soluble nitrogen method as previously described (Rutherfurd, 2010), and calculated by following formula: DH (%) = [content of amino nitrogen in supernatant (mg/ml)/total content of nitrogen in sample (mg/mL)] × 100%.
2.2. Peptide sequence identification
The peptide composition in MSOP was measured by MALDI-TOF/TOF 5800 (AB Sciex, Framingham, MA, USA) at Jiyun Biotechnology Co., Ltd. (Shanghai, China) as previously described (Lu et al., 2021). MS spectra were collected in a molecular mass range of 500–3,000 Da, the peak detection signal-to-noise ratio was set to 15, and MS/MS spectra were analyzed to identify peptides using the Mascot Server (Matrix Science Inc, Boston, MA, USA) with the NCBI nr database.
2.3. Molecular docking with Discovery Studio 2019
The molecular structures of peptides were built, and energy was minimized using Discovery Studio 2019. The X-ray structure of Keap1 (PDB ID: 5DAD) was retrieved from the Protein Data Bank (https://www.rcsb.org), the water molecules, native ligands and heteroatom molecules were removed by Discovery Studio 2019, and PyRx 0.9 software was used to minimize the energy of the protein. Then, the CDOCKER module in Discovery Studio 2019 was selected for molecular docking using the default settings as previously described (Fan et al., 2022).
2.4. Cell culture and UVB irradiation
The HaCaT cell line was purchased from Beyotime Biotechnology (Shanghai, China), and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) (Hyclone) and 1% antibiotics (Hyclone) at 37 °C and 5% CO2 in a humidified incubator. HaCaT cells were seeded and adhered for 24 h. The cells were treated with various concentrations of MSOP and subsequently exposed to UVB radiation at a dose of 50 mJ/cm2 according to previous studies (Xiao et al., 2021). Cells without MSOP or UVB irradiation treatments were served as the control.
2.5. Cell viability assay
HaCaT cells (1 × 105) were seeded in 96-well culture plates and treated with various concentrations of MSOP for 24 h. Then, an MTT assay kit purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, Jiangsu, China) was used to measure the cell proliferation and cytotoxicity in response to MSOP treatments at different concentrations.
2.6. Antioxidant enzyme activities
MDA, SOD and GSH-Px activities were measured using colorimetric assay kits (Nanjing Jiancheng Bioengineering Institute) according to the manufacturer’s protocol.
2.7. Apoptosis detection
Cells were washed with PBS, stained with an Annexin V-FITC/PI kit (Beyotime Biotechnology), and measured with a CytoFLEX Flow Cytometer (Beckman Coulter, Inc. Brea, CA, USA). In addition, the apoptotic rate was analyzed by Modfit LT 6.0 software (www.vsh.com).
2.8. Quantitative proteomic analysis
According to the previous method (Miles et al., 2023), the cell pellet was extracted with lysis buffer. The protein extract was centrifuged at 15,000 g at 4 °C for 20 min, the supernatant was treated with acetone to precipitate protein, and NH4HCO3 solution (40 mM) was added to the pellets. The protein concentration was measured by a BCA protein assay kit (Nanjing Jiancheng Bioengineering Institute). Then, the proteins were reduced and alkylated before digestion by trypsin (Promega Corporation, Madison, WI, USA). The digestion was stopped by formic acid, and the samples were then vacuum-dried.
Digested peptides were separated with chromatography by an Easy-nLC1000 system (Thermo Fisher Scientific, Am Kalkberg, Osterode, Germany) as previously described (J. Guo et al., 2017). Separated peptides were detected in the Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) with a Michrom captive spray nanoelectrospray ionization (NSI) source. Spectra were scanned over the m/z range 300–4,000 Da at 120,000 resolution.
The raw data were further examined by PEAKS software (version 7.5, Bioinformatics Solutions, Waterloo, Canada) as previously described and a label-free approach was performed to detect the relative quantification by the Q module in PEAKS (Yang et al., 2023). Proteins were considered to be significant when p < 0.05 and a fold change of ≥2.
Bioinformatics analysis was performed according to a previous method (J. Liu et al., 2015). Functional annotation was performed by Blast2GO, (version 6.0.1) and protein pathway analysis was performed by the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg).
2.9. Quantitative real-time PCR (qRT-PCR) analysis
As previously described (Huang et al., 2021), total RNA was isolated and purified by the TransZol Up Plus RNA kit (Beijing TransGen Biotech Co., Ltd., Beijing, China), cDNA was reverse transcribed by TransScript® All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (Beijing TransGen Biotech Co., Ltd), and the primers used in this study are presented in Table S1. The mRNA level of the target gene was calculated by the 2-ΔΔCT method and normalized to the β-actin level.
2.10. Statistical analysis
All data are presented as the mean ± standard deviation (SD). The normally distributed data were assessed by ANOVA followed by Tukey’s post hoc test (SPSS, version 19.0, Chicago, IL, USA), and data that did not meet the assumptions of ANOVA were analyzed by the Mann-Whitney test (MATLAB R2012a, Natick, MA, USA). Statistical significance was defined as p < 0.05.
| 3. Results | ▴Top |
3.1. Optimization of MSOP preparation
In single-factor experiments, the results showed that the optimum hydrolysis conditions of time, temperature and E/S ratio were 3.5–4.5 h, 25–35 °C and 0.5–1.5%, respectively (Figure S1a–c). In addition, the response surface experiment showed that DH (%) = 87.75 − 0.58A − 1.31B + 0.080C + 0.020AB + 0.33AC + 0.55BC − 6.52A2 − 5.41B2 − 5.01C2 (Where A is the time; B the temperature; C the amount of enzyme), and the most suitable variables for the time, temperature and E/S ratio were 4.31 h, 32.30 °C and 1.25%, respectively. (Figure S1d and Table S2–3). At that optimized condition, the DH reached 83.71%, while the predicted DH was 80.24%.
3.2. Composition of MSOP
The compositions of peptides in MSOP were determined by MALDI-TOF/TOF. The results of the first-order mass spectrometry showed that the majority of the parent ions were less than m/z 1,000 (Figure S2a). Three peptides were found to be dominant in the MSOP, with molecular weights of 506.2860 Da, 524.2478 Da and 524.2478 Da, respectively (Figure S2b–d). Further analysis by second-order mass spectrometry identified the amino acid sequences of these three peptides, which were VADML (Val-Ala-Asp-Met-Leu), IARF (Ile-Ala-Arg-Phe) and SSPSF (Ser-Ser-Pro-Ser-Phe)
3.3. Predicted interactions between three peptides and Keap1
In this study, a total of 44 target pharmacophores were predicted to interact with three peptides. We compared the fit value of peptides with those of targets, and the targets with high fit values are listed in Table 1. The results showed that VADML, IARF and SSPSF interacted with Keap1 (4zy3 and 5dad), playing a vital role in the Keap1/Nrf2-ARE pathway involved in oxidative stress regulation. Therefore, Keap1 was used as the receptor for subsequent molecular docking.
 Click to view | Table 1. The potential targets of three peptides with high fit value screened by Discovery Studio 2019 |
The interactions between the peptides and Keap1 were characterized with ECI, EVDW and RMS Gradient. As shown in Table 2, the -ECI values of VADML, IARF and SSPSF were 92.3, 73.6 and 63.0, respectively, while the corresponding -EVDW values were 10.1, 5.6 and 3.7. According to Figure 1a, there are seven amino acid residues of Keap1 that interact with VADML. Among them, ArgB:233 formed attractive charges, SerB:231 and GlnB:276 formed conventional hydrogen bonds, TrpB:232, SerB:239 and ProB:235 formed carbon hydrogen bonds, and TyrB:230 formed pi-alkyl groups. There are nine amino acid residues involved in the interactions between IARF and Keap1 (Figure 1b). Among them, AspB:278 and AspB:279 formed attractive charges, SerB:231, ArgB:233, TrpB:240, SerB:275 and GlnB:276 formed conventional hydrogen bonds, and ProB:235 and SerB:275 produced carbon hydrogen bonds. There are 7 seven amino acid residues involved in SSPSF interactions with Keap1 (Figure 1c). GlnB:276, SerB:231, SerB:234 and TrpB:240 formed conventional hydrogen bonds, TrpB:232 formed carbon hydrogen bonds and TrpB:230 and PheB:282 formed pi-alkyl bonds.
 Click to view | Table 2. The molecular docking results of three peptides with Keap1 |
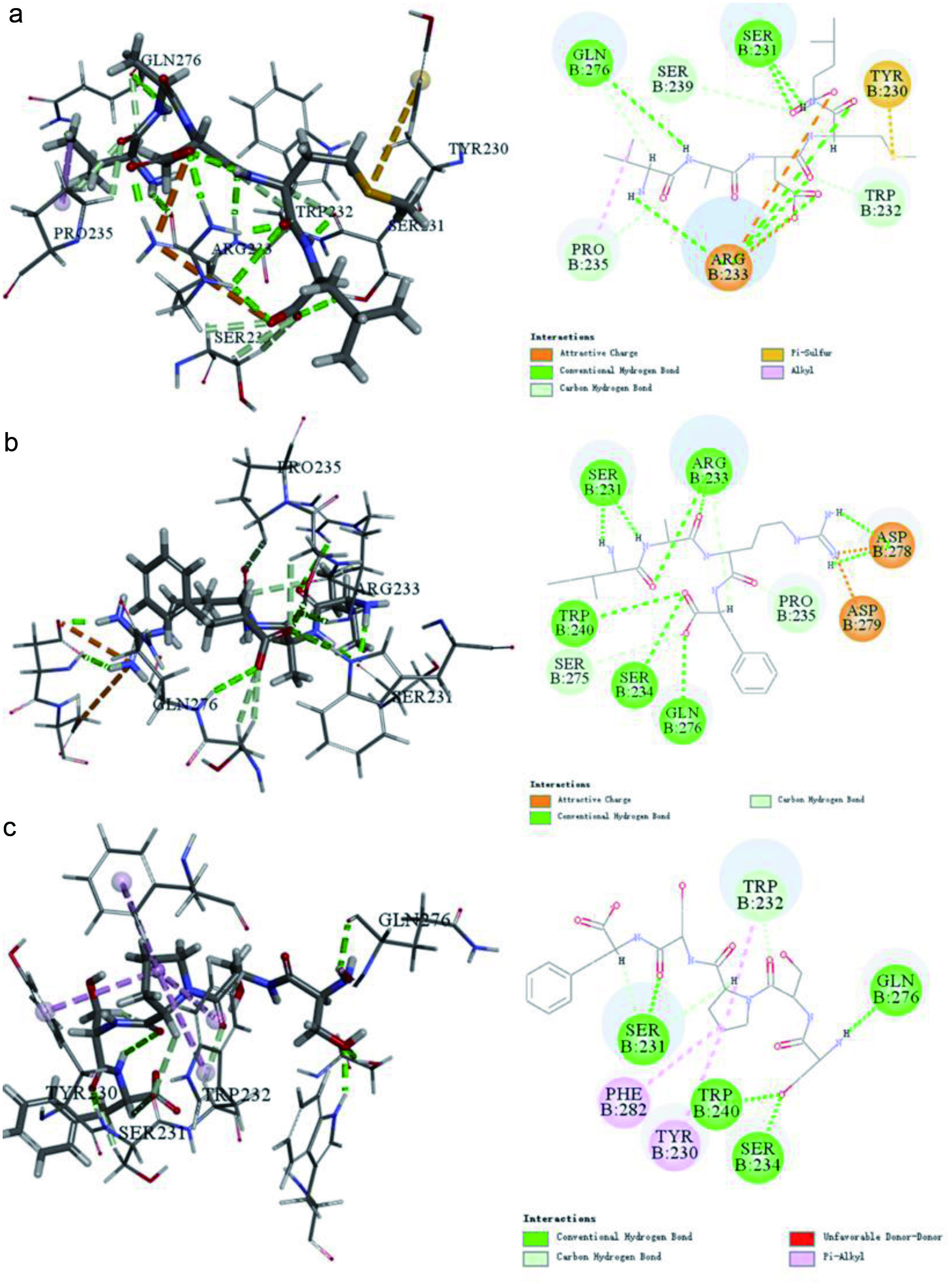 Click for large image | Figure 1. Interactions between Keap1 and peptide predicated Discovery Studio 2019. (a) VADML, (b) IARF, (c) SSPSF. |
3.4. Protective effects of MSOP against UVB-induced injury in HaCaT cells
We investigated the effect of MSOP on the proliferation of HaCaT cells after exposure to UVB. Cell viability was reduced to 57.37% by UVB irradiation in the absence of MSOP but significantly restored with MSOP treatment, and the highest cellular survival rate was observed in the high dosage MSOP-treated group (Figure 2a).
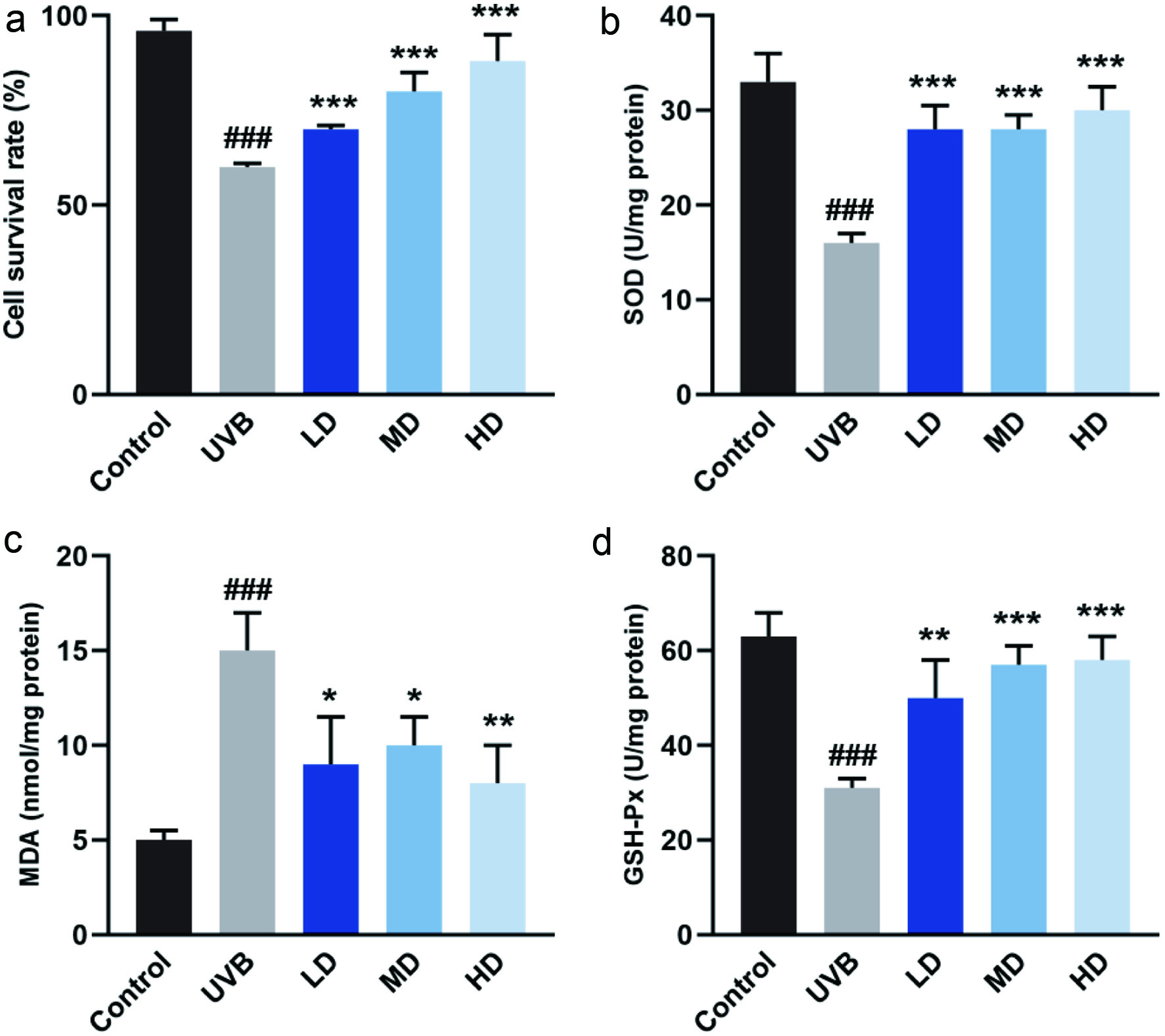 Click for large image | Figure 2. Antioxidant activity of MSOP in human keratinocytes HaCaT cells exposed to UVB. (a) Cell viability in response to UVB and MSOP treatments. (b) activity of SOD. (c) Content of MDA. (d) Activity of GSH-Px. ### p < 0.001 compared to control group. *p < 0.05, **p < 0.01 and ***p < 0.001 compared to UVB group. |
3.5. Antioxidant effects of MSOP in UVB-induced HaCaT cells
To investigate whether the radical scavenging activity of MSOP was mediated by antioxidant enzymes, the activities of antioxidant enzymes were examined in HaCaT cells after UVB exposure. SOD and GSH-Px activity were enhanced by MSOP treatments in a dose-dependent manner. Furthermore, in comparison with the control group, the level of MDA in the UVB-induced group was increased, but treatment with MSOP decreased the level of MDA (Figure 2b–d).
3.6. Protective effects of MSOP against UVB-induced apoptosis
The lower left quadrant represents viable and healthy cells, which were decreased by UVB treatment, but restored after MSOP treatment at a high dosage. The right quadrant represents the apoptotic cells, including early and late apoptotic cells, which decreased from 21.3% to 16.2% in MSOP-treated cells (Figure 3).
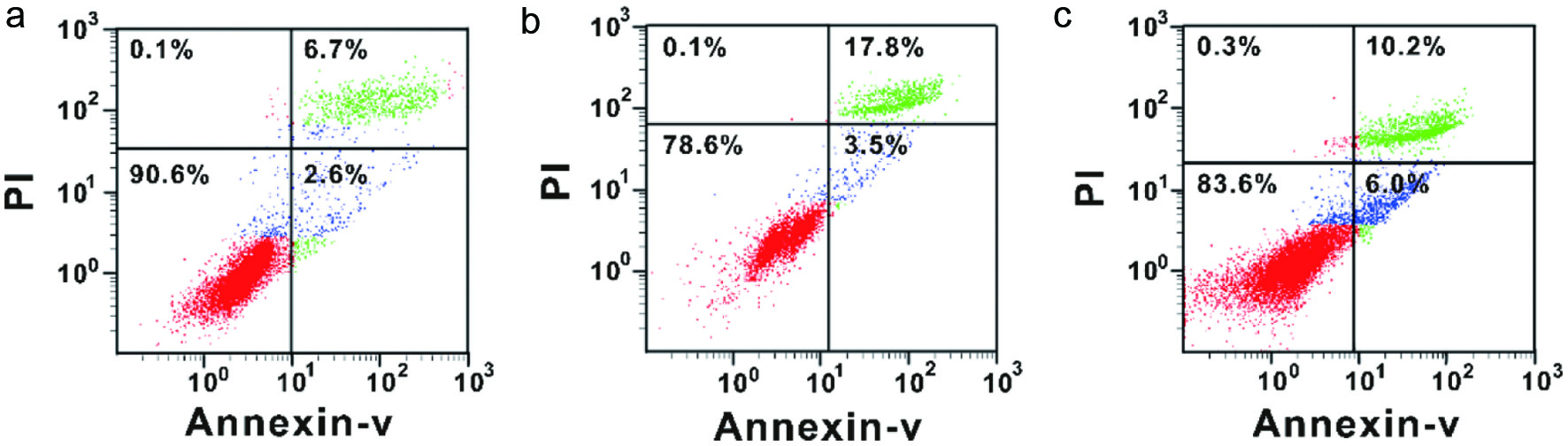 Click for large image | Figure 3. The anti-apoptosis effects of MSOP measured by flow cytometric analysis. (a) Control group. (b) UVB group. (d) High-dosage UVB group. |
3.7. Analysis of differentially expressed proteins
Figure 4a shows the overlap of differentially expressed proteins (DEPs) (fold change ≥2 or ≤0.5) from the control, LD, MD and HD groups compared to the UVB group. The numbers of DEPs in the control, LD, MD, and HD groups were 286, 204, 176 and 157, respectively. Among them, 119 proteins were upregulated and 167 were downregulated. The numbers of DEPs upregulated in LD, MD, and HD were 98, 104, and 79, respectively, while the corresponding numbers for downregulation were 106, 72 and 78. In addition, 23 DEPs were identified in all pairwise comparisons (Figure 4b and Table S4).
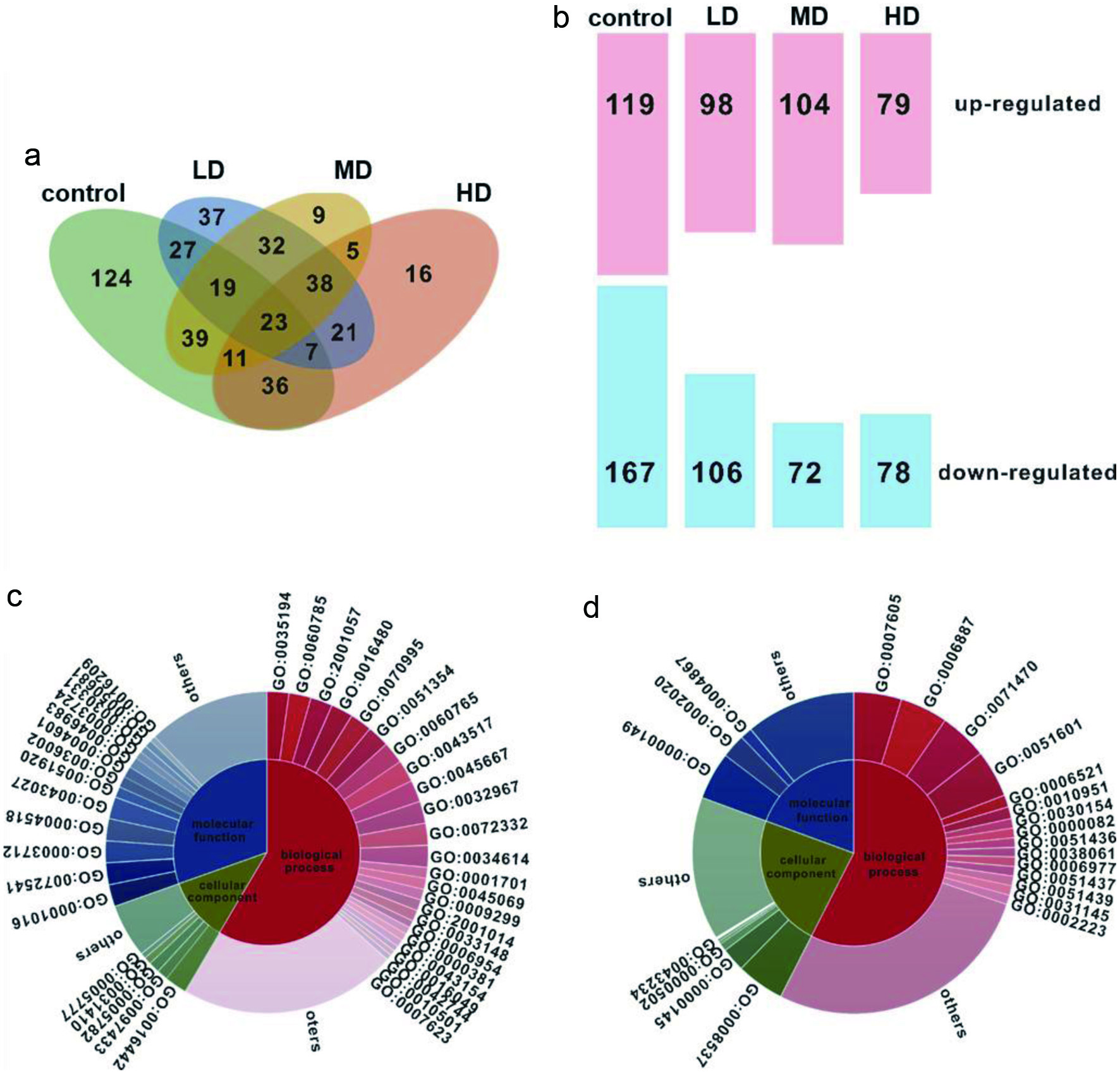 Click for large image | Figure 4. Quantitative proteome analysis in HaCaT cells in response to UVB and MSOP treatments. (a) Venn diagram of differentially expressed proteins (DEPs) (fold change ≥2 or ≤0.5) in response to UVB and MSOP treatments. (b) Numbers of upregulated and downregulated proteins in HaCaT cells. Upregulated (c) and downregulated (d) DEPs categorized by biological process, molecular function, and cellular component. |
To validate the accuracy of the proteomic data, the mRNA levels of 12 DEPs were assessed by qRT-PCR. Figure 5 shows that seven DEPs (qcr2, calX, uba1, dhe3, lap2B, ehd2 and nrf2) were upregulated, and the remaining five DEPs (6 pgd, serA, keaP1, lamP1 and ero1A) were downregulated. The Pearson correlation coefficients for proteomics and qRT-PCR data were ≥ 0.9.
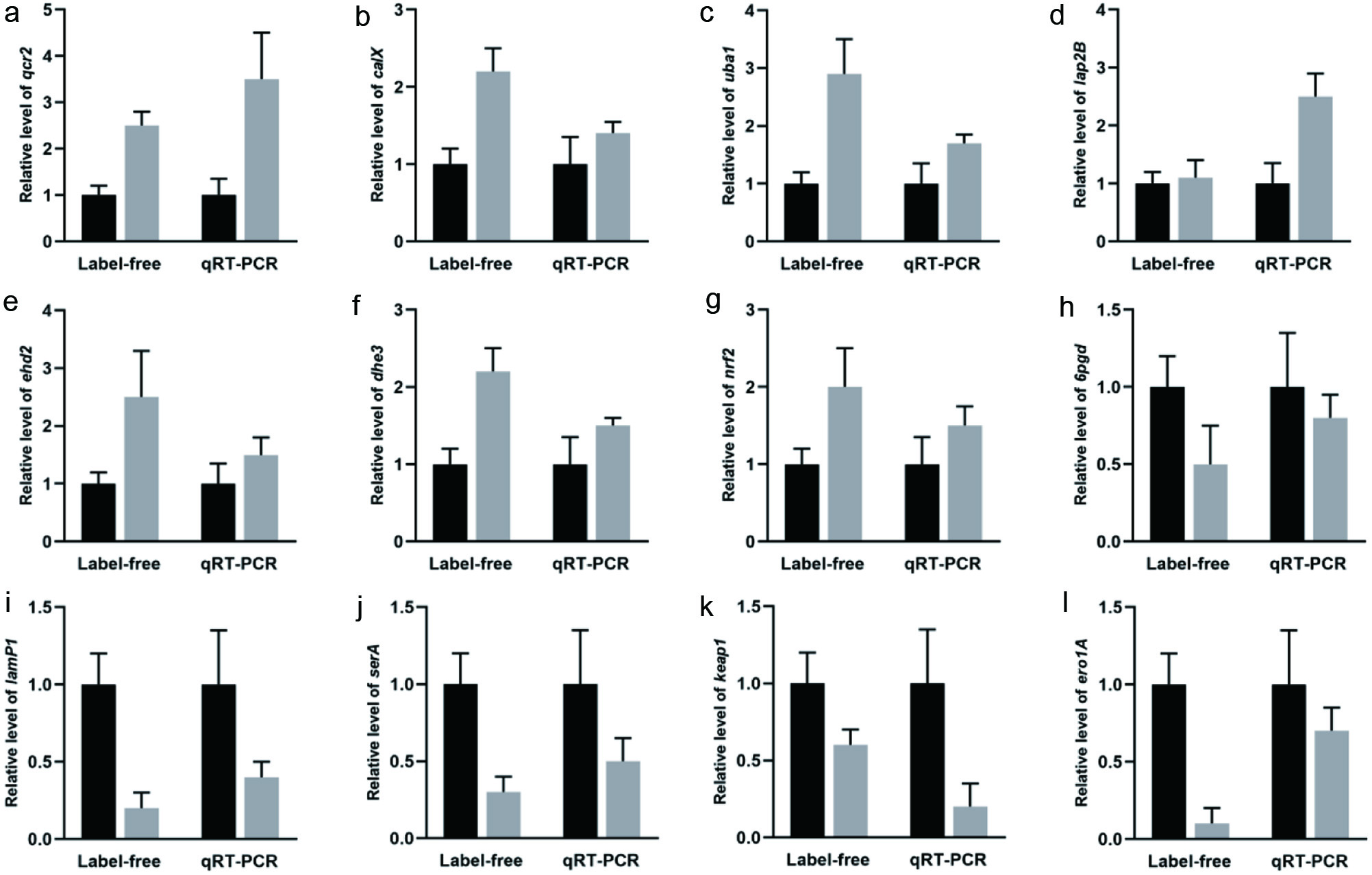 Click for large image | Figure 5. Expression of differentially expressed proteins measured by quantitative proteome and qRT-PCR in HaCaT cells. (a) qcr2, (b) calX, (c) uba1, (d) dhe3, (e) lap2B, (f) ehd2, (g) nrf2, (g) 6pgd, (i) serA, (j) keap1, (k) lamP1, (l) ero1A. |
3.8. Functional analysis of DEPs
The proteins with increased expression were categorized as biological processes (BP), cellular components (CC), or molecular functions (MF). In the comparison of UVB and HD, the proteins with increased expression in the BP category were posttranscriptional gene silencing by RNA, regulation of apoptosis involved in tissue homeostasis, reactive nitrogen species metabolic process, etc. The proteins in the CC category were RISC complex, dense body and peroxisomal matrix. The proteins in the MF category were RNA polymerase III regulatory region DNA binding, peroxynitrite reductase activity, transcription cofactor activity, etc (Figure 4c). The proteins with decreased expression in the BP category were defense response to sensory perception of sound, exocytosis, cellular response to osmotic stress, etc. The proteins in the CC category were proteasome activator complex and exocyst, while the proteins in the MF category were SNARE binding, protease binding, serine-type endopeptidase inhibitor activity, etc (Figure 4d). In addition, the results from KEGG differential pathway analysis showed that the DEPs were involved in 9 metabolic pathways that were mainly related to the oxidative stress pathway (Table S5).
| 4. Discussion | ▴Top |
Biologically active peptides are inactive within the sequence of their parent protein and can be released by enzymatic hydrolysis either during gastrointestinal digestion or food processing (Dhar et al., 2023; Ucak et al., 2021). In this study, MSOP was prepared via pepsin, the major components were identified to be VADML, IARF and SSPSF, and the potential antioxidant activity was predicted by Discovery Studio 2019. In addition, a UVB-induced HaCaT cell model was established to explore the anti-photoaging and antioxidant activity of MSOP in vitro, and this cell model was widely used in evaluating the anti-photoaging activity of Salvia plebeia R. Br ethanol extract, bioactive peptides from Skipjack Tuna cardiac arterial bulbs and flexible liposomes co-loaded with apigenin and doxycycline as previously described (Guo et al., 2023; Kong et al., 2023; Liu et al., 2023)
Discovery Studio is a powerful research platform for the life sciences that integrates a comprehensive suite of informatics, modeling and simulation solutions (Han et al., 2020). From the results of reverse docking, we found that all three peptides interacted with Keap1 (4zy3 and 5dad) (Table 1). Nuclear factor erythroid 2-like 2 (Nrf2), is a transcription factor that serves as a key regulator of the cellular response to oxidative stress. Under normal physiological conditions, Keap1 maintains low levels of Nrf2 by promoting its Cul3-rbx1-mediated ubiquitination and subsequent proteasomal degradation. Under stressful conditions, ROS modify sensor cysteine residues within Keap1 and block the degradation of Nrf2. Newly synthesized Nrf2 then translocates to the nucleus, binds to antioxidant response elements, and increases the transcription of numerous genes that encode cellular defense and repair enzymes (Ong et al., 2023; Suzuki et al., 2023). This coordinated and rapid upregulation of multiple genes ensures a robust response to many types of cellular stress (Dai et al., 2023). The results of molecular docking showed that VADML, IARF and SSPSF interacted with Keap1 and showed potential Keap1 inhibitory activity (Figure 1). Thus, we proposed a tentative inference that MSOP played an antioxidative role by regulating the Keap1-Nrf2/ARE signaling pathway.
Superoxide radicals are formed in all living cells exposed to oxygen, by various biochemical systems in the cells. SOD catalyzes dismutation of the superoxide anion (O2-) into hydrogen peroxide, which protects organisms against the toxic effects of superoxide radicals (Khayatan et al., 2023). GSH-Px is located primarily in the cytosol and has a general specificity in detoxifying H2O2 and converting lipid hydroperoxides to nontoxic alcohols (Brigelius-Flohé and Flohé, 2020). MDA can exacerbate the actions of superoxide ions by impairing endothelium dependent relaxation and propagation of lipid peroxidation by chain reaction in membranes (Del Rio et al., 2005). In our experiment, MSOP treatment dose-dependently increased the activities of SOD and GSH-Px, and decreased the content of MDA (Figure 2), which activated the defensive mechanism against ROS attack in the living HaCaT cells.
However, the underlying mechanism mechanism of MSOP activity in the UVB-irradiated HaCaT cellular model is extremely complex, and it remains unclear which proteins are involved in the beneficial effects of MSOP. Therefore, quantitative proteome technology was used in this study. Research has shown that that the Keap1/Nrf2/ARE signaling pathway plays a significant role in protecting cells from oxidative stress (Hassanein et al., 2020; Shi et al., 2021; Yuan et al., 2020). Under quiescent conditions, the transcription factor Nrf2 interacts with the actin-anchored protein Keap1, which is largely localized in the cytoplasm. This quenching interaction maintains low basal expression of Nrf2-regulated genes. However, upon UVB- irradiated photodamage, Nrf2 is released from Keap1, escapes proteasomal degradation, translocates to the nucleus, and transactivates the expression of several dozen cytoprotective genes that enhance cell survival (Figure 6). The present study showed that the MSOP caused significant upregulation of Nrf2 and downregulation of Keap1 in HaCaT cells, which may contribute to the beneficial effects of MSOP in a UVB-induced HaCaT cellular model.
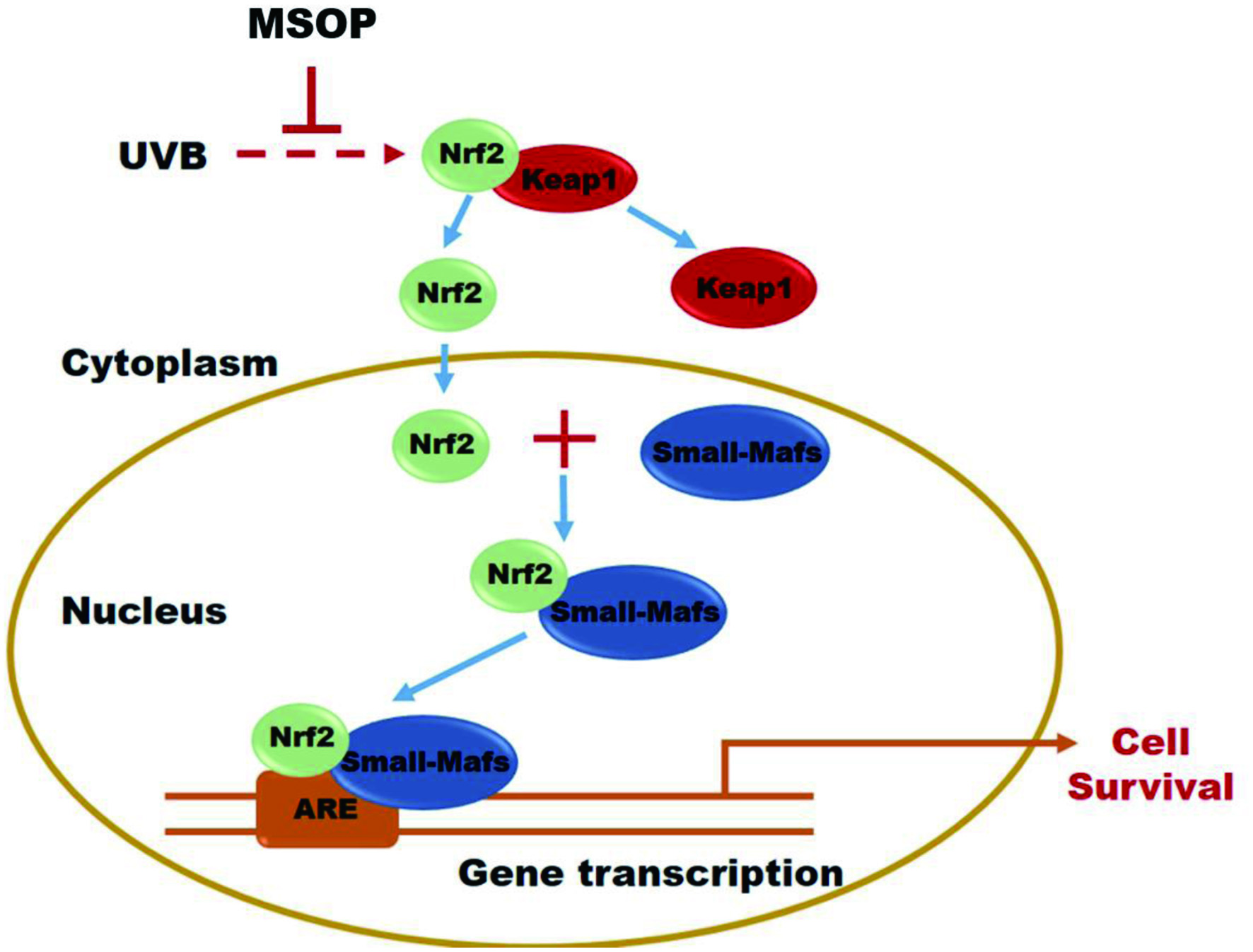 Click for large image | Figure 6. General scheme for the beneficial effects of MSOP through Keap1/Nrf2/ARE signaling pathway regulation. |
In conclusion, we optimized the preparation method of MSOP with an 80.24% degree of hydrolysis and identified three peptides (VADML, IARF and SSPSF), which were highly abundant and predicted to show potential antioxidant activity due to their interactions with Keap1 via Discovery Studio 2019 simulation. In addition, the antioxidant activity of MSOP was investigated in vitro via a UVB-induced HaCaT cell injury model, and the underlying mechanism was explored via quantitative proteomics, especially on the regarding of the Keap1/Nrf2/ARE pathway. MSOP might offer an alternative for protecting skin against UVB exposure.
| Supplementary material | ▴Top |
Table S1. Primers used in this study for qRT-PCR.
Table S2. Central composite design quadratic polynomial model, experimental data and actual values.
Table S3. ANOVA for the quadratic polynomial model.
Table S4. Twenty-three DEPs were identified in all comparisons with the UVB group.
Table S5. KEGG pathway analysis of DEPs.
Figure S1. Optimization of enzymatic hydrolysis conditions. Effect of time (a), temperature (b) and E/S ratio (c) on the enzymatic treatment. (d) 3D and 2D response surfaces demonstrating the effects of enzymolysis conditions.
Figure S2. Identification of the molecular mass and amino acid sequence of the peptides from MSOP. (a) Full MS scan. MS/MS spectrum of the ion 506.2860 m/z (b), 524.2478 m/z (c) and 548.1384 m/z (d).
Figure S3. Cell survival rate of HaCaT cells at different concentrations of MSOP.
Acknowledgments
This work was sponsored by Regional Demonstration Project of Marine Economic Innovation and Development in 2014 and 2016, the National Key Research and Development Program of China (2018YFD0901102), and K.C. Wong Magna Fund of Ningbo University.
Conflict of interest
The authors report no conflict of interest.
| References | ▴Top |