| Journal of Food Bioactives, ISSN 2637-8752 print, 2637-8779 online |
| Journal website www.isnff-jfb.com |
Original Research
Volume 25, March 2024, pages 13-24
Exploring the phytochemical composition and pharmacological effects of fermented turmeric using the isolated strain Lactobacillus rhamnosus FN7
Kai-Jiun Loa, #, Sandeep Choudharya, #, Chi-Tang Hob, Min-Hsiung Pana, *
aInstitute of Food Science and Technology, National Taiwan University, Taipei 10617, Taiwan
bDepartment of Food Science, Rutgers University, New Brunswick, NJ, 08901, USA
#These authors contributed equally.
*Corresponding author: Min-Hsiung Pan, Institute of Food Science and Technology, National Taiwan University, No.1, Section 4, Roosevelt Road, Taipei 10617, Taiwan. Tel: +886-2-33664133; Fax: +886-2-33661771; E-mail: mhpan@ntu.edu.tw
DOI: 10.31665/JFB.2024.18368
Received: December 28, 2023
Revised received & accepted: January 26, 2024
| Abstract | Top |
Turmeric (Curcuma longa), widely used in Asia as a spice, preservative, and colorant, contains curcuminoids known for diverse pharmacological benefits, including antimicrobial properties. However, their hydrophobic nature hampers bioavailability. Addressing this, we hypothesized that Lactic Acid Bacteria (LAB) fermentation could enhance curcuminoid content and bioactivity. This study isolated LAB strains to ferment turmeric and investigated the phytochemical and pharmacological outcomes. Twelve LAB strains from various sources were tested for fermenting 3% turmeric in MRS broth. L. rhamnosus FN7 emerged as a robust strain, tolerating turmerics antibacterial properties and increasing curcuminoid content and anti-inflammatory effects. Fermented turmeric exhibited higher phenolic and flavonoid contents and improved radical scavenging activity than its non-fermented counterpart. Additionally, L. rhamnosus FN7 survived under simulated gastrointestinal conditions, indicating probiotic potential. Our findings suggest that L. rhamnosus FN7 fermentation significantly boosts turmerics biochemical attributes, positioning it as a promising functional food.
Keywords: Fermented turmeric; Curcuminoids; Lactic acid bacteria; L. rhamnosus
| 1. Introduction | Top |
Turmeric, a member of the Zingiberaceae family, is derived from the rhizome of Curcuma longa which is extensively cultivated in India, China, and various South Asian countries (Li et al., 2011). It has a rich history of traditional medicinal use, serving as both the natural coloring and flavoring agent in food (Prasad and Aggarwal, 2011). Previous research has highlighted the diverse health benefits of turmeric, encompassing anti-inflammatory, antineoplastic, hypolipidemic, and other pharmacological activities (Sharifi-Rad et al., 2020). In contemporary times, turmeric has gained widespread popularity as one of the most sought-after medicinal herbs, spices, and functional dietary supplements. The biological activity of turmeric is predominantly attributed to its abundant array of bioactive compounds, including curcuminoids, monoterpenes, sesquiterpenes, diterpenes, triterpenoids, alkaloids, and sterols (Li et al., 2011). Among these constituents, curcuminoids, which constitute 5% of the total weight in turmeric, have garnered significant global scientific attention for their pharmacological and therapeutic properties (Sharifi-Rad et al., 2020).
Within turmeric, curcuminoids exist three notable analogs including curcumin (diferuloylmethane), demethoxycurcumin (DMC), and bisdemethoxycurcumin (BDMC). These compounds manifest distinctions in their methoxy substitutions on the aromatic ring. Curcumin features two symmetric o-methoxy phenols connected by the , -unsaturated -diketone moiety. In contrast, BDMC, also symmetric, lacks two o-methoxy substitutions, and DMC displays an asymmetric structure with one phenyl ring incorporating o-methoxy substitution. Among these curcuminoids, curcumin predominates in turmeric, succeeded by DMC and BDMC (Jayaprakasha et al., 2002). A commercially available curcumin mixture comprises 77% curcumin, 17% DMC, and 3% BDMC (Anand et al., 2008). So far, extensive in vitro research indicates the antioxidant, cardioprotective, anti-inflammatory, anti-cancer, anti-Alzheimer, anti-diabetic, anti-microbial activities, and other medicinal properties of curcuminoids (Sharifi-Rad et al., 2020). However, despite their diverse bioactivities, the hydrophobic nature of dietary turmeric curcuminoids poses a limitation, rendering them practically insoluble in water. Additionally, the curcuminoid constituents of turmeric exhibit low absorption, rapid metabolism, and elimination in the body, thereby restricting their bioavailability when orally consumed as a supplement (Anand et al., 2007). To tackle these challenges, increasing the curcuminoid content in turmeric products or modifying curcuminoids to enhance their bioavailability presents a potential solution.
Fermentation has played a crucial role in human food production and consumption for an extended period. The process involves the breakdown of complex organic substances using microorganisms to enhance various aspects of food, including vitamins, essential amino acids, anti-nutrients, proteins, appearance, flavor, and aroma (Sharma et al., 2020). The United States Food and Drug Administration (USFDA) recognizes certain microorganisms as safe for producing fermented foods, such as Aspergillus oryzae and Penicillium roqueforti for koji and cheese, Saccharomyces cerevisiae for bread, and lactic acid bacteria (LAB) for yogurt and pickles. The market offers a variety of fermented food products based on the selection of microorganisms, raw materials, and manufacturing techniques (Anal, 2019). Recently, several reports have shown that the fermentation of turmeric by Generally Recognized as Safe (GRAS) microorganisms can enhance its efficacy. For instance, Trichoderma spp. fungus can boost the antioxidant and antibacterial activity of non-fermented turmeric, while fermented Aspergillus oryzae turmeric improves protective effects on CCl4-induced liver damage in rats (Mohamed et al., 2016). The solid-state fermentation of turmeric using Rhizopus oligosporus increases major curcuminoids, total flavonoids, and antioxidant activity (Lim et al., 2022). Similarly, turmeric fermented with L. fermentum increases total curcumin content, exhibiting higher anti-inflammatory activity compared to unfermented turmeric (Sharma et al., 2022; Yong et al., 2019). Likewise, L. johnsonii IDCC 9203 fermented turmeric strongly inhibits pro-inflammatory cytokines, enhances water solubility of major curcuminoids, and improves consumer acceptance compared to non-fermented turmeric (Kim et al., 2011). These findings suggest that the fermentation of turmeric using microorganisms could serve as an alternative approach to enhance the overall efficacy of turmeric.
To date, a few specific strains of LAB that exhibit effectiveness on turmeric have been documented. However, the impact of a diverse array of LAB strains on turmeric remains unexplored. Furthermore, the identification of a potential strain within diverse LAB strains that can withstand the antimicrobial effects of turmeric is very crucial. In addition, there are still unanswered questions regarding the effects of LAB fermentation on the phytochemical changes in turmeric, particularly in terms of its anti-inflammatory properties. Thus, the study aimed to address these gaps through LAB isolation from different sources and comparing their ability to ferment turmeric. The results revealed that turmeric fermented with the L. rhamnosus FN7 strain displayed a notable capability to increase curcuminoid content and overall phytochemicals in turmeric, thereby enhancing its anti-inflammatory activity. Furthermore, L. rhamnosus FN7 exhibited potential probiotic properties. These findings suggest that the utilization of Lactobacillus fermentation in turmeric has the potential to elevate its curcuminoid content and phytochemical properties, positioning fermented turmeric as a promising functional food for addressing inflammation.
| 2. Material and Methods | Top |
2.1. Sample preparation and LABs isolation
The turmeric powder utilized in the study was supplied by ASAKUSA AGRICULTURE PROCESSING CO., located in Hualien City, Taiwan. The powder underwent a cool-air drying process and was vacuum-sealed for storage at 20C. For LAB isolation, strains were isolated from distinct sources which included healthy human faeces, soil from turmeric cultivation (Hualien City, Taiwan), and a yogurt product from Uni-president in Taiwan. Briefly, each sample was mixed with 0.85% NaCl distilled water at a ratio of 1:10 and vortexed (Scientific Industries, USA) for 30 seconds. Subsequently, the mixture was suspended in sterilized 0.85% NaCl distilled water to prepare serial dilutions and 100 L of each dilution were evenly spread on the surface of De Man-Rogosa-Sharpe (MRS) agar plates containing 3% turmeric powder (TMRS) using a sterilized L-shaped glass spreader. The plates were then anaerobically incubated with oxygen adsorbent packs (Mitsubishi Gas Chemical Co., Japan) for 4872 hours at 37C. Following incubation, an individual colony from each source on the MRS agar plate was selected, and a new series of dilutions was prepared. The colonies were streaked in a zig-zag pattern on TMRS agar. The isolation process was repeated until pure colonies were observed on an agar plate. Finally, the putative LAB colony from the agar plate was cultured in MRS broth at 37C for 20 hours to determine through microscopic examination whether it was a single strain.
2.2. 16S rRNA gene sequencing for phylogenetic analysis of the isolated strains
The genomic DNA extraction was performed using the phenol/chloroform extraction method. After the centrifugation at 15,000g for one minute, cells were collected, and 600 L of phenol/chloroform/isopropanol (25:24:1) was added, and mixed well. Then the mixture was centrifuged at 15,000g for 5 minutes and the upper aqueous phase was carefully transferred to a fresh tube. The step was repeated two times. Subsequently, an equal volume of chloroform was added to mix with the aqueous layer, and centrifugated at 15,000g for 5 minutes. The resulting aqueous layer was again transferred to a fresh tube and four times the volume of absolute ethanol was added for DNA precipitation at 20C for one hour. DNA pellets were obtained by centrifugation at 15,000g for 15 minutes and washed twice with 70% ethanol at the same condition. DNA pellets were briefly air-dried and resuspended in TE buffer to obtain a DNA sample. The qualification was performed using the NANODROP 1000 spectrophotometer (Thermo Scientific, Taiwan) at 260 nm and 280 nm. The DNA samples prepared were used for PCR reactions and sequencing analysis.
The PCR reactions were done to amplify16S rRNA using universal primers, 27F (5-AGAGTTTGATCCTGGCTCAG-3) and 1492R (5-GGTTACCTTGTTACGACTT-3). The reactions were performed under the following conditions: 94C for 2 minutes; 35 cycles of 30 seconds at 94C, 30 seconds at 60C, and 2 minutes at 72C; a final extension at 72C for 5 minutes. The PCR products were purified using a DNA Clean & Concentrator-5 kit (Zymo Research, USA) and sequenced by the Center for Biotechnology at National Taiwan University.
Individual 16S rRNA sequences were compared against the NCBI non-redundant nucleotide database using the Basic Local Alignment Search Tool (BLAST). For phylogenetic analysis, the sequences of type strains most similar to the isolates were downloaded from the NCBI database and aligned using BioEdit with the ClustalW multiple alignment program under default settings. The MEGA11 software was utilized to construct the topological tree of LAB strains using the maximum likelihood program with the general time-reversible model and gamma-distributed with the invariant model. One thousand resamplings were employed to evaluate the level of support for the internal branches.
2.3. Bacterial culture conditions and turmeric fermentation
The bacterial culture medium employed for turmeric fermentation was MRS powder (NEOGEN, Lansing, MI). A single activated bacterial colony was picked up and transferred to a glass tube containing 3 mL of MRS medium, then incubated at 37C for 20 hours. Subsequently, 10% of the culture broth was transferred to fresh MRS broth and incubated at 37C for another 20 hours. For turmeric fermentation, 1% of the bacterial cultures grown for 20 hours (adjusted OD600 value to 1.0) in the log phase were inoculated into the MRS medium containing 3% turmeric and incubated under static conditions at 37C for 72 hours. Bacterial broth samples were collected at 12 hours intervals up to 72 hours. Colony-forming units were used to measure the growth of bacteria on the MRS agar plates. All experiments were performed with three biological replications.
2.4. Quantification of curcuminoids by High-performance Liquid Chromatography (HPLC) analysis
For HPLC analysis, curcuminoids were extracted from the freeze-dried fermented turmeric with little modifications from the previous study. Briefly, fermented turmeric was mixed with 1 mL of ethyl acetate containing 10 g/mL methyl red (internal control) and vortexed vigorously. Next, the supernatant was collected by centrifugation at 15,000g for 5 minutes, and the residual pellets were repeatedly extracted until became colorless. All of the supernatants were collected together and dried using nitrogen gas at 25C. Finally, the residue was suspended in acetonitrile and filtered with a 0.22 m syringe filter to obtain curcuminoid extract.
HPLC analysis was according to the previous study and performed by Jasco Pu-2080 plus an Intelligent HPLC pump system coupled with a UV-vis detector (set at 425 nm). The chromatographic separation was achieved using the C18 column (150 4.6 mm, 5 m; Agilent Technologies, USA) at 30C. The mobile phase was 0.1% formic acid in deionized water (A) and acetonitrile (B) at a flow rate of 1 mL/min. The elution gradient was used as follows: 60% A and 40% B in the beginning, decreased to 36% A at 7 minutes, maintained for 3 minutes, decreased to 10% A at 15 minutes, and returned to the original ratio at 17 minutes. The content of curcuminoids in sample extracts was calculated using the standard curve of pure curcumin, BDMC, and DMC.
2.5. Phytochemical properties of Isolated L. rhamnosus FN7 fermented turmeric
For sample preparation, 50 mg samples from both fermented and unfermented turmeric powders were taken and suspended in 1 mL of 70% aqueous ethanol, then stirred at 50C for 2 hours to extract the bioactive compounds. Subsequently, the supernatant was collected using centrifugation at 3,000g for 5 minutes and lyophilized. Finally, the residues were redissolved in methanol and filtered through a 0.22 m filter membrane for further use. The total phenolic content was measured by the Folin-Ciocalteau (FC) method with slight modification. In short, 12.5 L of the sample and 50 L of distilled H2O were mixed and added to a 96-well plate. Subsequently, 12.5 L of FC reagent was added, and the mixture was allowed to mix for 5 minutes. Finally, 125 L of 7% Na2CO3 was added and shaken for 90 minutes. The absorbance was measured at 750 nm against water blank (Synergy Instrument Inc., Vermont, USA). Gallic acid was served as the standard. For flavonoid content, an Aluminium chloride (AlCl3) colorimetric assay with slight modification was used. Briefly, 15 L of sample extracts and 45 L of methanol were added to a 96-well plate, along with 3 L of AlCl3 and 3 L of 0.1 M potassium acetate. Subsequently, 84 L of distilled H2O was added and incubated for 30 minutes. The absorbance of the acid-stable complex was measured at 415 nm against blank water (Synergy HT, BioTek Instrument Inc., Vermont, USA). Quercetin was applied as the standard. The DPPH, a stable radical was used to measure the total antioxidant potential of both fermented and unfermented turmeric samples. Briefly, 180 L of 0.1 mM DPPH in methanol was added into a 96-well plate, followed by 20 L of sample extracts. Further, the plate was incubated in the dark for 30 minutes and absorbance was measured at 517 nm. Ascorbic acid was used as standard.
2.6. Anti-inflammatory activity of fermented turmeric in vitro
To investigate the anti-inflammatory activity of fermented turmeric, RAW264.7 murine macrophages (derived from the American Type Culture Collection, Rockville, MD, USA) were selected in the study. RAW264.7 cells were seeded at a density of 1 106 cells/well in 24-well plates containing 1 mL of Dulbeccos modified Eagle medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 100 g/mL streptomycin, and 100 g/mL penicillin. The cells were cultured in a humidified atmosphere with 5% CO2 at 37C for 12 hours. Following, the medium was removed, and cells were gently washed twice with phosphate-buffered saline (PBS). Subsequently, the cells were incubated in FBS-phenol red-free DMEM containing four mM glutamic acid, then treated with various concentrations (final concentrations were 50, 100, 200, 250, 300, 350, and 400 g/mL) of both unfermented and fermented turmeric powder for 24 hours with lipopolysaccharide (LPS) at a final concentration of 100 ng/mL. The cell viability was analyzed by using a classical MTT assay. The Griess assay was used to evaluate the nitrite production in RAW264.7 macrophages.
2.7. Probiotic characteristics of isolated L. rhamnosus FN7 strain
The simulation of gastric fluid tolerance in isolated strain through different pH values. Briefly, 2% (v/v) overnight cultured bacterial broth was incubated in an MRS medium with different pH values (2.0, 2.5, and 3.0) under anaerobic at 37C. Bacterial broth samples were collected at 3 hours intervals up to 6 hours. Colony-forming units were used to measure the growth of bacteria on MRS agar medium, while the samples taken at 0 hours were used as a control. To determine bile salt tolerance, 2% (v/v) overnight cultured L. rhamnosus FN7 isolate was added into 6 mL of fresh MRS medium containing 0.1, 0.2, 0.3, 0.4, 0.5, and 1% of bile salts (Sigma-Aldrich, New Zealand) under anaerobic conditions at 37C. Bacterial broth samples were collected at 3 hours intervals up to 6 hours, and colony-forming units were used to measure the growth of bacteria on MRS agar medium. To analyze the milk fermentation capacity, one hundred milliliter aliquots of 10% (w/v) skim milk media (Millipore, Switzerland) was pasteurized at 85C for 30 minutes and cooled down to 40C. Subsequently, 2% (v/v) of L. rhamnosus FN7 strain was inoculated and incubated at 37C for 24 hours under anaerobic conditions. After incubation, the pH of cultured milk was measured.
2.8. Statistical analysis
The results are expressed as the mean SD from three separate trials. Statistical distinctions among groups were assessed using one-way ANOVA followed by students t-test analysis. A p-value <0.05 was considered indicative of a statistically significant difference between each group.
| 3. Result and discussion | Top |
3.1. 16S rRNA phylogeny of isolated strains
LAB strains are often associated with dairy products, human oral cavities and intestines, fecal matter, and compost. To isolate LAB strains suitable for the fermentation of turmeric, we selected soil from turmeric fields, commercial yogurt, and the gut microbiome of an Indian adult as potential sources for performing the isolation. Around 14 cultures were isolated from different sources that, include ten from human faeces (FN1, FN3, FN4, FN6, FN7, FN8, FN9, FN11, FN13, and FN14), three from yogurt (YN2, YN5, and YN10), and one from soil (SN12). As the morphological structure of microbes provides a better understanding of microbial physiology and allows us to identify them by species, we observed the morphology of all isolated strains. All the isolates were found to be rod-shaped except isolate FN6, which was cocci-shaped, consistent with the morphotype in a majority of LAB (Axelsson, 2004). Moreover, most of the isolates were Gram-positive bacteria except isolates FN6 and SN12. According to the previous reports, Lactobacillales are an order of gram-positive, thereby deducing that both strains (FN6 and SN12) may not belong to LAB. We further used 16S rRNA sequencing to identify these isolates. The result of the phylogenetic tree showed that the most of isolates from the human faeces have high identical similarities to the species L. rhamnosus (similarity >99%), including FN1, FN3, FN4, FN7, FN8, FN9, and FN11; they were grouped into the same clades (bootstrap support of 100%). The isolated strains from the yogurt were placed in the same group with L. casei ATCC393 (bootstrap support of 95%). However, two isolates (FN6 and SN12) from faeces and soil were grouped outside the Lactobacillus spp., and clustered to Enterococcus faecalis and Klebsiella variicola (Figure 1). Both E. faecalis and K. variicola are pathogens causing infection in humans and animals (Horsley et al., 2013; Martnez-Romero et al., 2018). Therefore, both these pathogenic strains were not introduced in turmeric fermentation.
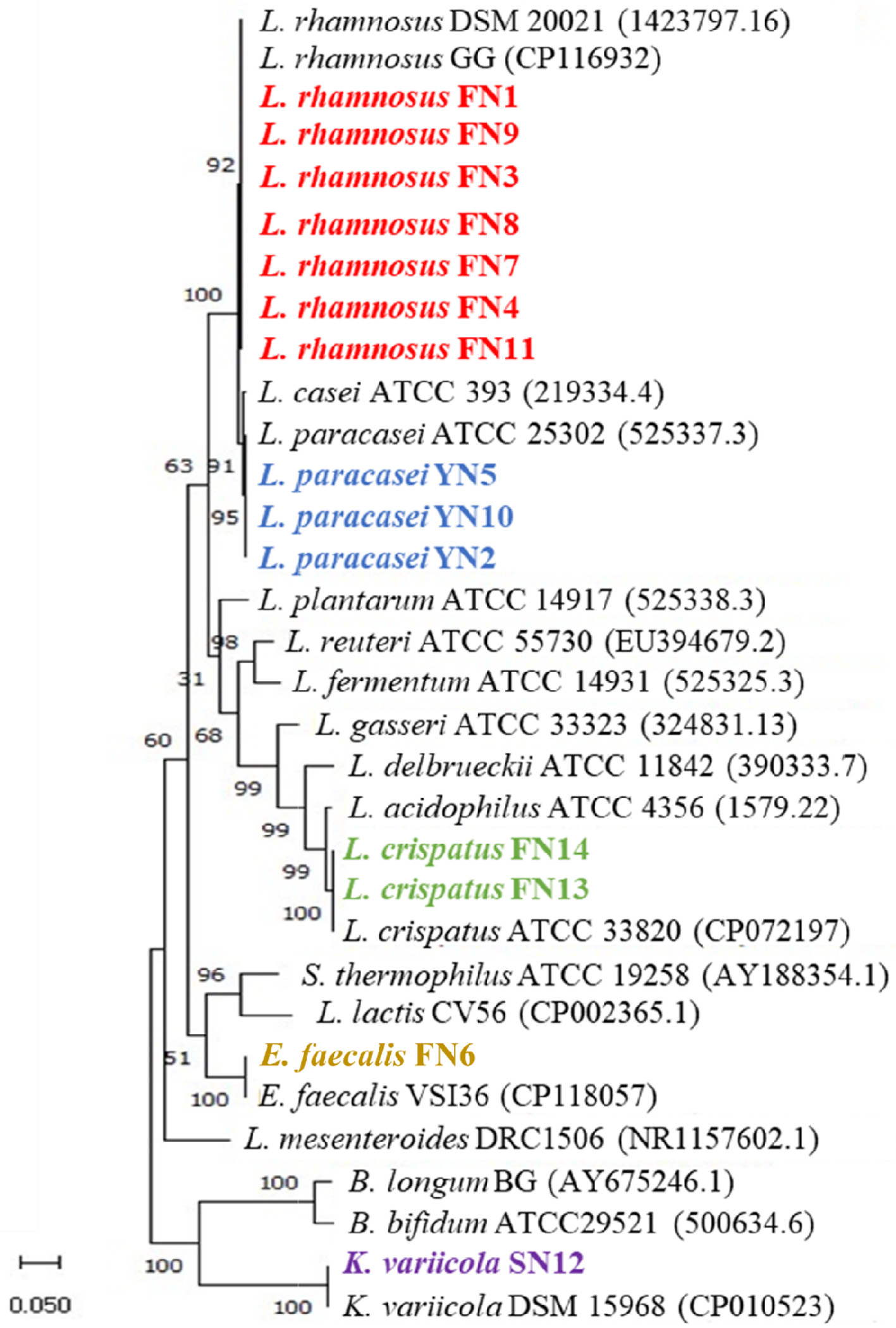 Click for large image |
Figure 1. 16S rRNA gene-based phylogenetic tree of the isolated strains. The tree was constructed using the maximum likelihood method. The bootstrap value expressed as a percentage of 1,000 replicates is given at each node. Nucleotide sequence accession numbers are indicated in parentheses. |
3.2. Isolated strains present the potential ability to ferment turmeric
To investigate whether the isolates present a potential ability to ferment turmeric, isolated LAB strains were introduced in MRS medium containing 3% turmeric to check the growth. In our study, results demonstrated that most of isolated strains unexpectedly encountered growth inhibition after inoculation which might be attributed to the antimicrobial properties of curcuminoids found in turmeric. Nevertheless, despite encountering suppression in their growth during the initial phase, these bacterial strains exhibit a sustained, albeit gradual, and growth rate (Figure 2). Remarkably, some strains even demonstrated a capacity to proliferate to levels equivalent to the control group in the later stages of the fermentation process, such as L. rhamnosus FN1, L. rhamnosus FN7, and L. rhamnosus FN8 (Figure 2a, f, and g). The enhanced resilience can be linked to the strains prior adaptation to external stress conditions within their specific sources, resulting in their ability to adapt more rapidly to the novel conditions presented by turmeric. Due to the fact that these strains still exhibit a certain level of growth ability during the fermentation of turmeric, all the isolated Lactobacilli strains were initially identified as potential strains capable of withstanding the curcuminoid effects of 3% turmeric.
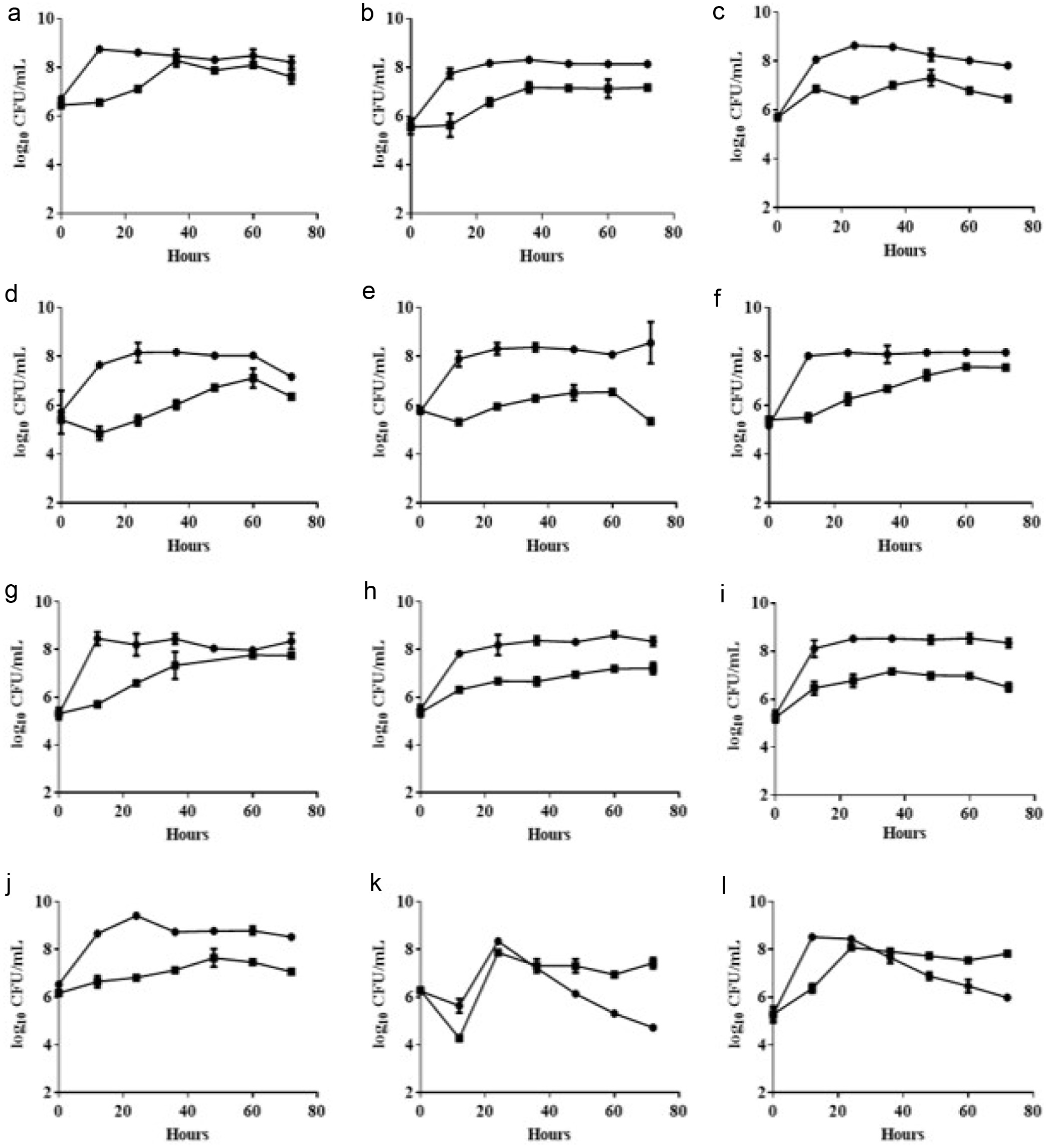 Click for large image |
Figure 2. Growth curve of isolated LAB strains in 3% turmeric conditions. MRS medium (circle) and MRS medium containing 3% turmeric (square). (a) L. rhamnosus FN1 (b) L. paracasei YN2 (c) L. rhamnosus FN3 (d) L. rhamnosus FN4 (e) L. paracasei YN5 (f) L. rhamnosus FN7 (g) L. rhamnosus FN8 (h) L. rhamnosus FN9 (i) L. paracasei YN10 (j) L. rhamnosus FN11 (k) L. crispatus FN13 (l) L. crispatus FN14. The 1% of bacterial broth (OD600 = 1.0) was inoculated into the MRS medium containing 3% turmeric powder within the static culture under 37C for 72 hours. The bacterial growth was evaluated using viable colonies forming unit (CFU). All experiments were carried out in triplicate, and bar values are expressed as the mean standard deviation. |
It is noteworthy that two isolated strains, L. crispatus FN13 and L. crispatus FN14, exhibit a significant decrease in biomass after 24 hours of cultivation under MRS medium. However, the addition of turmeric appears to sustain their growth (Figure 2k and l). Intriguingly, both strains belong to the L. crispatus species. These findings suggest that L. crispatus may recognize components within turmeric as nutritional sources, thereby supporting their growth and metabolic activities. To elucidate whether the strain is a critical factor for the survival of strains during turmeric fermentation, we conducted additional experiments using various standard strains. Our experimental results revealed that the majority of strains were significantly inhibited in media containing turmeric. Only a limited number of strains were able to survive, including B. longum BCRC 11847, L. gasseri BCRC 14619, L. reuteri BCRC 14625, L. rhamnosus BCRC 10940, L. rhamnosus GG BCRC 16000, and L. fermentum BCRC 12190 (Figure S1). The behavior of these specific strains could be attributed to the presence of specific cell surface components and genes that enable bacteria to withstand the antimicrobial effects of curcuminoids found in turmeric (Marathe et al., 2010). These results may also explain why the variety of strains we selected is not extensive (Figure 1). Furthermore, it also indicates that, despite certain strains being evolutionarily closely related, there are still significant differences in their characteristics of turmeric fermentation.
3.3. Curcuminoid content of turmeric increases after fermentation using isolated LAB strains
Curcuminoids are the key bioactive compounds that contribute to the wide range of pharmacological activities of turmeric (Sharifi-Rad et al., 2020). Despite the numerous bioactivities, the use of curcuminoids has been limited due to their thermal degradation, photodegradation, oxidation, alkaline hydrolysis, and acid hydrolysis (Peram et al., 2017). Therefore, it was curious to investigate the effect of identified potential LAB strains on the curcuminoid content of fermented turmeric. The curcuminoid content of fermented samples were analyzed using HPLC. The results revealed that fermentation with Lactobacillus improved the curcuminoid content (Figure 3). After the fermentation of turmeric, the curcuminoid content was significantly improved by the L. rhamnosus FN7 strain. Meanwhile, the curcuminoid content after fermentation by other isolated strains was insignificant except for FN8, FN9, FN10, and FN11 isolates. A possible explanation for the curcuminoid increase in turmeric after fermentation could be the utilization of enzymes by LAB strains. As reported in the earlier study, microorganisms use enzymes such as cellulases to aid in the extraction process by breaking or hydrolyzing plant tissues (Rosenthal et al., 1996). Similar results were observed by Yong, during the fermentation of turmeric using lactic acid bacteria (Yong et al., 2019). Unfortunately, our result revealed that none of the LAB strains displayed cellulase activity in isolates, as illustrated by L. rhamnosus FN7 (Figure S2). It is worth noting that we also assessed the curcuminoid concentration in fermented turmeric using six different types of strains with the potential for turmeric fermentation (Figure S1). The results indicated that there was no significant increase in curcumin content after fermentation (Figure S3). These results indicate that not all strains are capable of surviving the fermentation process possessing the ability to increase the curcuminoid content.
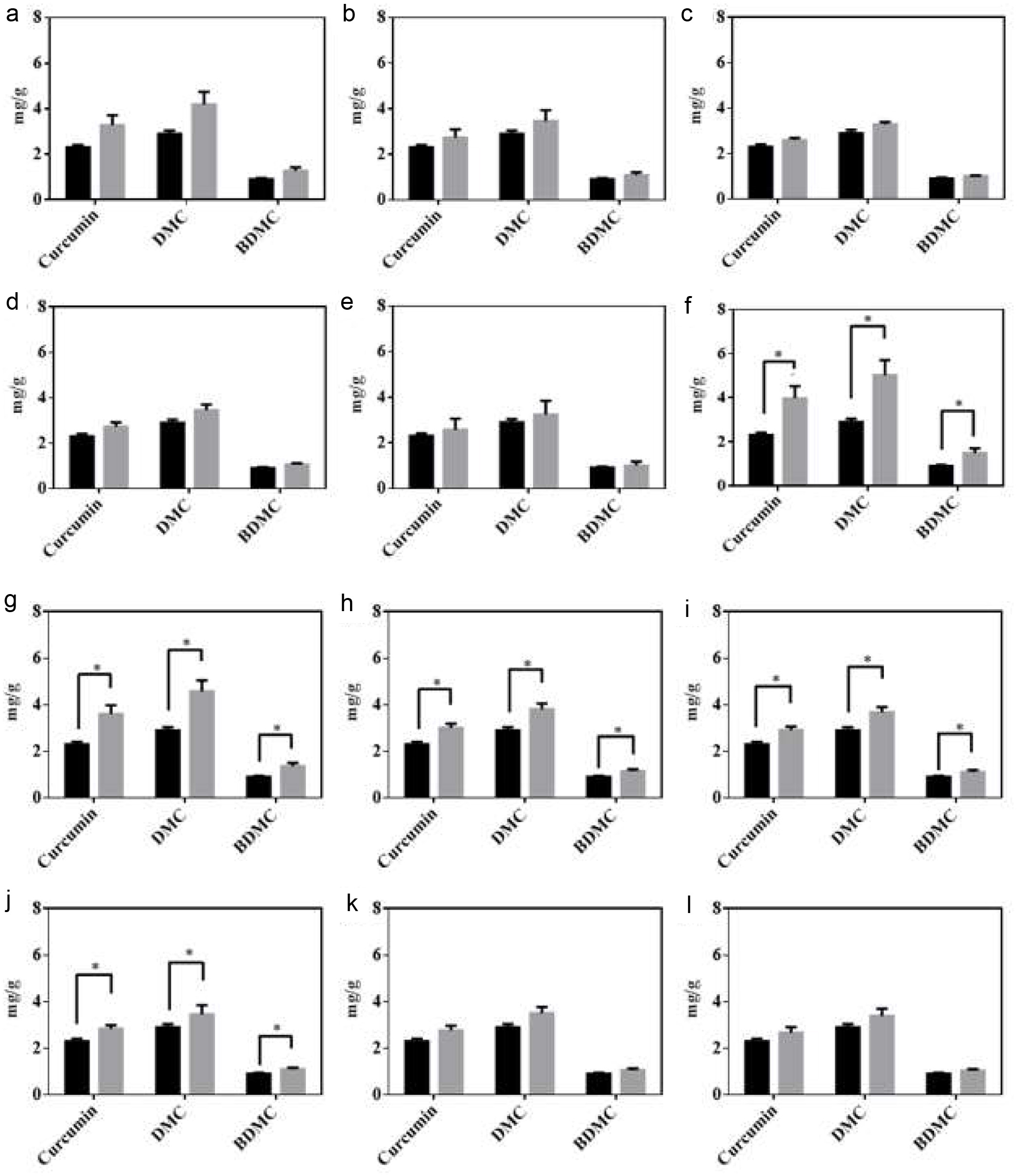 Click for large image |
Figure 3. HPLC analysis of curcuminoids in unfermented (black bar) and fermented turmeric (grey bar). (a) L. rhamnosus FN1 (b) L. paracasei YN2 (c) L. rhamnosus FN3 (d) L. rhamnosus FN4 (e) I L. paracasei YN5 (f) L. rhamnosus FN7 (g) L. rhamnosus FN8 (h) L. rhmanosus FN9 (i) L. paracasei YN10 (j) L. rhamnosus FN11 (k) L. crispatus FN13 (l) L. crispatus FN14. After fermentation, curcuminoids were extracted from the freeze-dried fermented turmeric powders and analyzed using the HPLC system comprised of a C18 column with the UV-vis detector (set at 425 nm). All experiments were carried out in triplicate, and bar values are expressed as the mean standard deviation. Statistics were analyzed by unpaired t-test, *p < 0.05. |
To explore the potential mechanism of L. rhamnosus FN7 improving curcuminoid content, we further examined the interaction between varying percentages of bacterial inoculation in an MRS medium containing various concentrations of pure curcumin to ascertain whether LAB either uptake or binds curcumin to the cell wall components. As illustrated in Figure S4, it was observed that the increase in curcumin was directly proportional to the biomass of bacteria. The observation suggests that the peptidoglycan within the bacterial cell wall can bind to curcumin, which may contribute to part of the mechanism leading to an increase in curcuminoids during the fermentation of turmeric by L. rhamnosus FN7. Moreover, our findings are also consistent with previous studies (Mun et al., 2014).
On the other hand, it has been commonly seen that curcumin is present in higher concentrations than DMC and BDMC (Jayaprakasha et al., 2002). However, during HPLC quantification, an abnormal change in the ratio of curcuminoid content (especially Curcumin and DMC) of turmeric was observed (Figure 3). To further resolve this problem, an experiment was conducted to examine the impact of two different heating methods i.e. pasteurization and sterilization. The results, outlined in Figure S5, revealed that after the pasteurization, the concentration of curcumin and DMC was similar. Whereas, following the sterilization of turmeric, the concentration of the DMC was found to be higher than that of curcumin demonstrating its higher thermal stability than curcumin. There are two main possible reasons for curcumin decrease: (1) the thermal degradation of the -diketone bond in its structure, which is quite vulnerable to break at temperatures higher than 100C (Suresh et al., 2009). (2) The presence of additional functional methoxy groups in the curcumin structure lowers its stability (Heffernan et al., 2017).
3.4. L. rhamnosus FN7 fermentation enhances the anti-inflammatory activity of turmeric
LPS is an endotoxin derived from the outer cell wall of gram-negative bacteria, which plays a critical role in the regulation of inflammation (Yang et al., 2016). To assess the potential anti-inflammatory properties of unfermented and fermented turmeric, LPS-induced in vitro model was conducted. As turmeric fermented by L. rhamnosus, FN7 showed a higher curcuminoid than other groups. The turmeric sample derived from L. rhamnosus FN7 fermentation was selected as a target in our study. As shown in Figure 4a, it was found that the LPS reduced the cell viability of RAW264.7 cells more than 80% compared to the control group, while tested unfermented and fermented turmeric exhibited an influence on cell viability across all concentrations. The turmeric concentrations below 200 g/mL had a significant difference in the cell viability compared to the control. As the concentration of the treatment increased, the cell proliferation gradually increased from the lower concentration to the higher concentration in fermented turmeric compared to unfermented turmeric. The result demonstrated the stimulatory effects of fermented and unfermented turmeric on the growth of RAW264.7 cells. However, there was no significant difference between unfermented and fermented turmeric treatment upon cell viability.
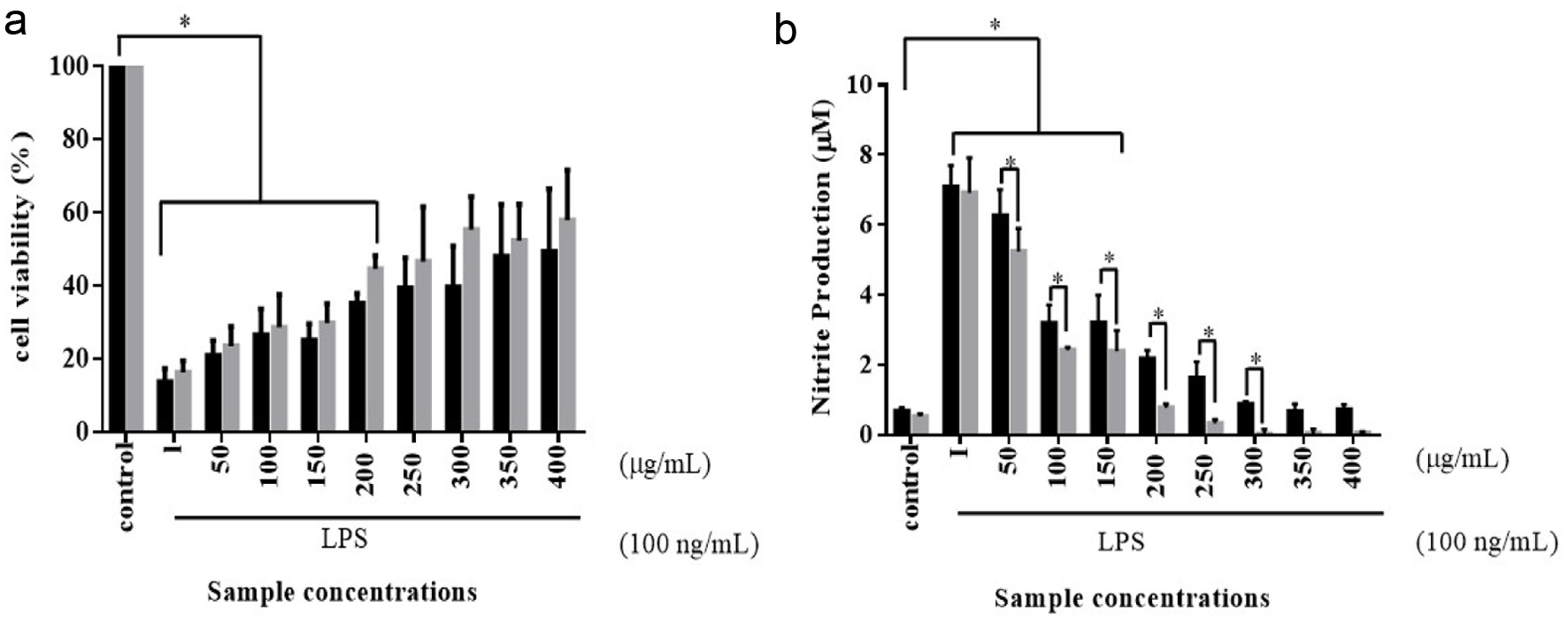 Click for large image |
Figure 4. Cell viability (a) and nitrite production (b) of RAW264.7 murine macrophages were treated with various concentrations of unfermented turmeric (black bar) and fermented turmeric (grey bar) for 24 hours. RAW264.7 murine macrophages were cultured with lipopolysaccharide (LPS) and treated with various unfermented and fermented turmeric powder for 24 hours under a humidified atmosphere with 5% CO2 at 37C. The cell viability was analyzed using a classical MTT assay, and the Griess assay was used to evaluate the nitrite production. Results were statistically analyzed with the LSD method. Data are present as mean SD from at least triplicate wells and three independent experiments. Statistics were analyzed by unpaired t-test, *p < 0.05. |
As mentioned in previous studies, a high level of NO could induce apoptosis directly or indirectly rendering many cells susceptible to apoptosis (Allione et al., 1999). In the current study, the accumulated nitrite in the cell culture supernatant was determined via Griess method as an index for NO produced by the LPS-induced RAW264.7 cells. To investigate the anti-inflammatory effect, we examined whether fermented turmeric could modulate nitrite (NO) synthesis in LPS-stimulated cultures of the RAW264.7 murine macrophages cells. As shown in the Figure 4b, it is evident that the production of NO in LPS-stimulated cells was significantly decreased when exposed to the fermented turmeric, starting from the concentrations of 50 g/mL to 300 g/mL, as compared to the unfermented turmeric, suggesting that the protective effect of fermented turmeric was attributed to the reduction of NO. While, concentrations beyond 300 g/mL, there was no significant difference in the inhibitory effect on nitrite production between the two types of turmeric. Therefore, the analysis of our data strongly suggests that fermented turmeric showed more significant reductions in LPS-induced NO production in comparison to unfermented turmeric. The result is also consistent with a recent study, suggesting the suppression of the JNK signal pathway by fermented turmeric results in a profound protective effect on the LPS-induced RAW264.7 cells (Yong et al., 2019).
3.5. L. rhamnosus FN7 fermentation improves the phytochemical properties of turmeric
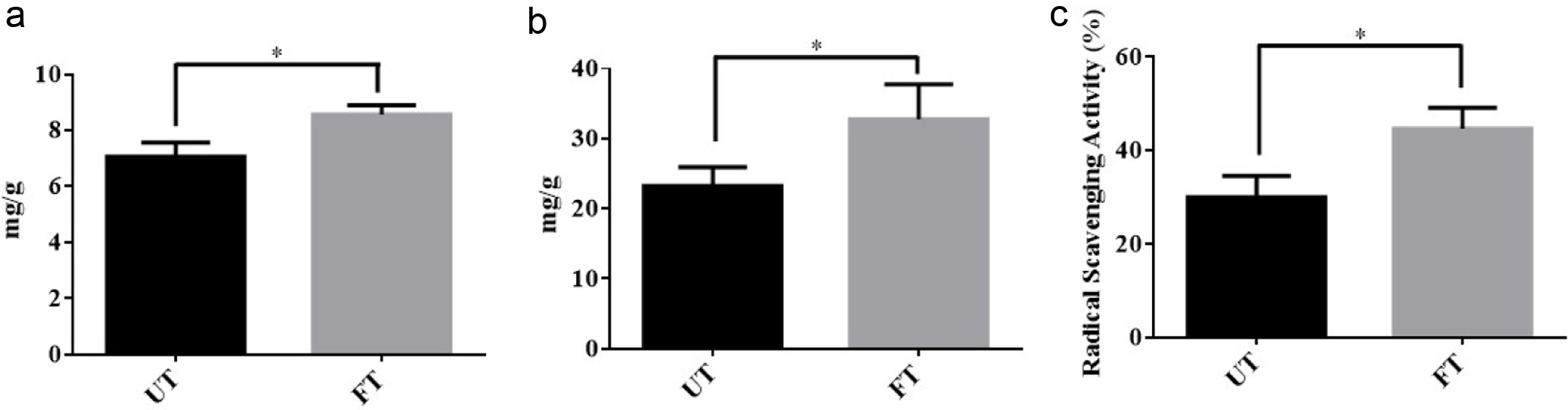
To date, around 235 compounds, primarily phenolic compounds and terpenoids, have been identified from the turmeric (Li et al., 2011). Due to their antioxidative properties of phenolic compounds, they are beneficial against diabetes, cardiovascular, mutagenesis, carcinogenesis, and neurogenerative disease. Thus, the content of phenolic compounds in fermented turmeric was disclosed before and after fermentation. As shown in Figure 5a, the results revealed a significant elevation in the phenolic compounds after fermentation. Numerically, the total phenolic compounds value of unfermented sample was 7.06 mg gallic acid/g which was significantly enhanced to 8.57 mg gallic acid/g after L. rhamnosus FN7 turmeric fermentation. The result was similar with previous research work in which phenolic compound content was investigated during the fermentation of the blueberry-carrot blend and mulberry juice using Lactobacillus strains. These two fermented products possess more phenolic compounds after fermentation (Kwaw et al., 2018; Mauro et al., 2016). The possible reasons for the rise in phenolic levels in fermented turmeric can be associated with the breakdown of glycosylated phenolics present in the unfermented turmeric matrix, resulting in the release of bound and insoluble phenolics from the cellular structures of plant material during the fermentation process (Mauro et al., 2016). Moreover, the existing research supports the notion that microorganisms can break down phenolic complexes into simpler and more bioavailable phenolic compounds that can be easily absorbed (Adetuyi and Ibrahim, 2014). Therefore, the conversion and depolymerization of complex compounds by LAB during fermentation could be a possible explanation for the rise in phenolic content.
 Click for large image |
Figure 5. Phytochemical property of fermented turmeric by L. rhamnosus FN7. (a) Total Phenolic Content (b) Total Flavonoid Content (c) Radical Scavenging Activity of unfermented turmeric (UT) and fermented turmeric (FT). Freeze-drying fermented and unfermented turmeric powders were used to extract the total Phenolic and Flavonoid compounds and quantification was performed with a slightly modified Folin-Ciocalteau (FC) method and Aluminium chloride (AlCl3) colorimetric assay, respectively. DPPH analysis was used to evaluate the Radical Scavenging Activity of fermented and unfermented turmeric. All experiments were carried out in triplicate, and bar values are expressed as the mean standard deviation. Statistics were analyzed by unpaired t-test, *p < 0.05. |
We further evaluated the total flavonoid content in the fermented turmeric and compared it with unfermented turmeric. The flavonoids are low molecular weight, secondary metabolites having a polyphenolic structure that not only play a role in determining the color and fragrance of flowers in fruits and vegetables but also have health-promoting effects (Panche et al., 2016). In turmeric, the flavonoids include not only curcuminoids but also other bioactive compounds such as luteolin-7-O-(6-p-hydroxybenzoyl--D-glucopyranoside), luteolin 7-O--D-glucopyranoside, apigenin-7-O--D-glucopyranoside, luteolin, apigenin, apigenin 7-O-rhamnoside 4-O-glucoside, 7-methoxyapigenin-6-C-glucoside (Shabana et al., 2015). As shown in Figure 5b, the total flavonoid contents of fermented turmeric showed a similar trend as total phenolic compounds. The total flavonoid content in fermented turmeric was determined significantly higher in comparison to unfermented turmeric. Before fermentation, the total flavonoid content value of the unfermented turmeric was 23.2 mg quercetin/g, which was significantly enhanced to 32.7 mg quercetin/g after L. rhamnosus FN7 fermentation. The increase in total flavonoid contents might be attributed to the enzymatic breakdown of complex polyphenols into simpler flavanol compounds during the fermentation process (Kwaw et al., 2018). Moreover, it may also be resulted from the higher curcuminoid content (Priyadarsini, 2013).
Reactive oxygen species (ROS) are molecules with unpaired electrons generated by various factors, including environmental pollutants, radiation, chemicals, toxins, physical stress, deep-fried and spicy foods. It has been reported that ROS significantly contributes to the onset of various physiological disorders, such as cellular damage, aging, cancer, and conditions affecting the liver, nervous system, heart, and kidneys (Madamanchi et al., 2005). To determine whether the scavenging ability of turmeric is enhanced after L. rhamnosus FN7 fermentation, the scavenging ability of DPPH radicals was investigated before and after fermentation. As shown in Figure 5c, the radical scavenging ability of fermented turmeric was significantly improved by L. rhamnosus FN7 fermentation. The radical scavenging ability of unfermented turmeric was 30.0 2.6% which significantly increased to 44.6 2.6% after fermentation of turmeric, indicating fermentation as the main cause of the enhancement in scavenging ability which may be due to the increase in total phenolic content (Ng et al., 2011). Moreover, it can also be attributed to the bacteria metabolization of the sugar attached to phenolic compounds and anthocyanins, leading to increased aglycone production, which enhances the radical scavenging capabilities of fermented turmeric (Sharma et al., 2023). In the past, it also has been reported that probiotic microorganisms chelate the metal ion and scavenge free radicals (Curiel et al., 2015). Overall, these findings suggested that turmeric fermented with L. rhamnosus FN7 helps in enhancing its phytochemical properties.
3.6. L. rhamnosus FN7 presents the potential ability to be applied as probiotics
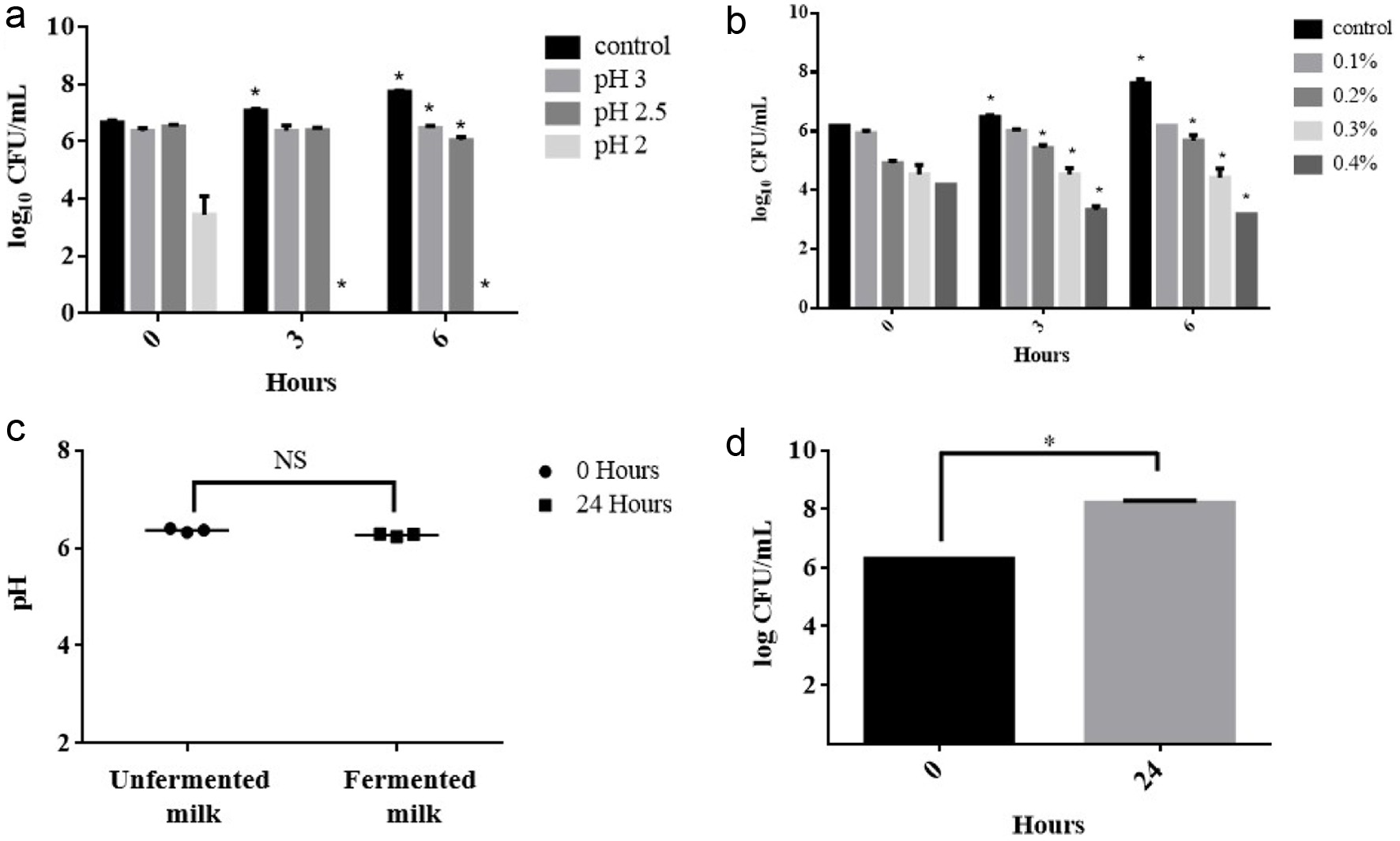
LAB to become probiotic, need to exhibit specific attributes, including the ability to withstand gastrointestinal conditions (such as low pH and high bile salt concentrations) (Collins et al., 1998). In past, Lactobacilli strains are highly recognized as promising probiotics, and are extensively employed in both food and non-food applications. In order to be used as a probiotic, the Lactobacilli strain must survive in the low pH conditions of gastric juice in the stomach (Goldin and Gorbach, 1992). In this study, to investigate whether L. rhamnosus FN7 can be employed as a potential probiotic, we further utilized in vitro assays to simulate and analyze the survival rate of L. rhamnosus FN7 in GI conditions. As shown in Figure 6a, at pH 3, the results revealed that the strain CFU/mL remained constant even after 6 hours. When exposed to pH 2.5, the CFU/mL of the L. rhamnosus FN7 showed a slight decrease after 6 hours of incubation. However, when the strain was introduced to pH 2, it was not able to survive and succumbed within hours of incubation. In addition, it is important to note that in the presence of food, the pH level typically rises around pH 3 which usually takes 2 to 4 hours for the stomach to empty after food ingestion (Erkkil and Petj, 2000). Therefore, such conditions can enhance the survivability of L. rhamnosus FN7 strain within the low pH of the human gastrointestinal tract.
 Click for large image |
Figure 6. The probiotic property assay of fermented turmeric by L. rhamnosus FN7. (a) pH tolerance, (b) Bile tolerance, (c and d) Milk fermentation capacity. For pH and bile tolerance, the L. rhamnosus FN7 strain was cultured in the MRS medium with different conditions under anaerobic at 37C for 6 hours. Milk fermentation capacity was performed by incubation of L. rhamnosus FN7 in 10% (w/v) skim milk. The viable colonies forming unit (CFU) was used to evaluate the growth of the L. rhamnosus FN7 strain in all experiments, and the pH of cultured milk was measured. Statistics were analyzed by unpaired t-test, *p < 0.05. |
Bile salts synthesized in the liver from cholesterol are known to create critical conditions for Lactobacillus strains to survive in the gastrointestinal tract (Erkkil and Petj, 2000). To address this, we investigated the bile tolerance ability of L. rhamnosus FN7 strain. Previous research has identified a concentration of 0.3% bile salts as a crucial threshold for screening bile-resistant strains (Gilliland et al., 1984). Similarly, Goldin and Gorbach suggested the same concentration level to select probiotics for human use (Goldin and Gorbach, 1992). In our test, the colony-forming units (CFU/mL) of L. rhamnosus FN7 were evaluated in bile salt concentrations of 0.1, 0.2, 0.3, 0.4, 0.5, and 1.0%. The results showed a gradual decrease in CFU/mL following inoculation, with successive colony counts at 3 hours intervals up to 6 hours (Figure 6b). Notably, no colony was observed at bile concentrations of 0.5% and 1%, respectively. Hence, the resistance of the L. rhamnosus FN7 strain to bile salts diminishes as the salt percentage increases. However, the strain can effectively survive at usual concentrations of bile salt in the gastrointestinal tract which typically encountered after food consumption (Erkkil and Petj, 2000).
Milk fermentation generally involves the conversion of lactose into lactic acid by LAB, particularly Lactobacilli and Lactococci, which leads in the reduction of the pH of the fermented milk (Surono and Hosono, 2011). To assess the milk fermentation capability and the ability of L. rhamnosus FN7 strain to digest lactose, an experiment was conducted. Thus, it was observed that there was negligible change in pH after 24 hours of incubation at 37C, while the strain CFU/mL increased from 6.3 to 8.2, suggesting that milk as a source of energy for the strain growth (Figure 6c and d). In addition, it is worth noting that various report indicates that L. rhamnosus GG strain cannot ferment lactose, which is consistent with our finding (Gorbach et al., 2017). However, a few strains of the same species have been reported to possess the capacity to ferment milk (Jeffrey et al., 2020). Hence, the probable reason for the L. rhamnosus FN7 inability to ferment the milk could be attributed to the frameshifts in the anti-terminator (lacT) and 6-phospho--galactosidase (lacG) genes, which affect the Lactobacillus strain ability to utilize D-lactose (Kankainen et al., 2009).
| 4. Conclusion | Top |
Recent research has increasingly focused on leveraging LAB fermentation to augment the bioactive functionality of foods. In this vein, our study has targeted the bioactivity enhancement of turmeric, a spice renowned for its health benefits but limited by the bioavailability of its active compounds, curcuminoids. Through meticulous strain selection, we have identified L. rhamnosus FN7 as a potent strain for turmeric fermentation, capable of augmenting its curcuminoid content. This process not only improves the phytochemical attributes of turmeric but also transforms it into a potent functional food with significant anti-inflammatory properties.
Our findings reveal that fermented turmeric, through the action of L. rhamnosus FN7, exhibits an increased concentration of curcuminoids and a broader spectrum of phytochemicals, thereby enhancing its efficacy in treating inflammatory conditions. Looking ahead, it is imperative to conduct thorough research to unravel the full spectrum of health benefits offered by L. rhamnosus FN7, particularly in its role as a probiotic for commercial application.
Moreover, our investigation has shed light on the mechanisms underlying the enhanced bioactivity post-fermentation. It has been elucidated that the peptidoglycan layer in the cell walls of Gram-positive bacteria such as Lactobacillus forms a complex with curcuminoids, facilitating their bioavailability. This binding phenomenon positions the Lactobacillus-curcuminoid complex as a promising natural vehicle for disease prevention, offering a novel approach to utilizing turmeric and its derivatives in therapeutic applications. The potential of such a natural carrier holds immense promise, warranting further exploration into its role in disease management and prevention strategies
| Supplementary Material | Top |
Suppl 1. Growth curve of different BCRC LAB strains in 3% turmeric conditions. MRS medium (circle) and MRS medium containing 3% turmeric (square). (a) L. gasseri BCRC 14619, (b) L. rhamnosus GG BCRC 16000, (c) Lactobacillus sp. MCL40.
Suppl 2. Cellulase activity test of L. rhamnosus FN7.
Suppl 3. Quantification of curcuminoids in unfermented (black bar) and fermented turmeric (grey bar) by different BCRC LAB strains. (a) B. longum BCRC 11847 (b) L. rhamnosus GG BCRC 16000 (c) L. gasseri BCRC 14619 (d) L. reuteri BCRC 14625 (e) L. rhamnosus BCRC 10940 (f) L. fermentum BCRC 12190.
Suppl 4. Effect of various percentage of L. rhamnosus FN7 biomass cell wall binding property in curcumin medium.
Suppl 5. Comparison of curcuminoids change in unfermented turmeric (UT) during Post-Heat retention time after autoclave.
Acknowledgments
We would like to acknowledge the service provided by the DNA Sequencing Core of the Center for Biotechnology, National Taiwan University.
This work was supported by the Ministry of Science and Technology, Taiwan [109-2320-B-002 -012 -MY3 and 110-2320-B-002 -019 -MY3].
Conflict of interest
The authors declare no conflict of interest.
| References | Top |