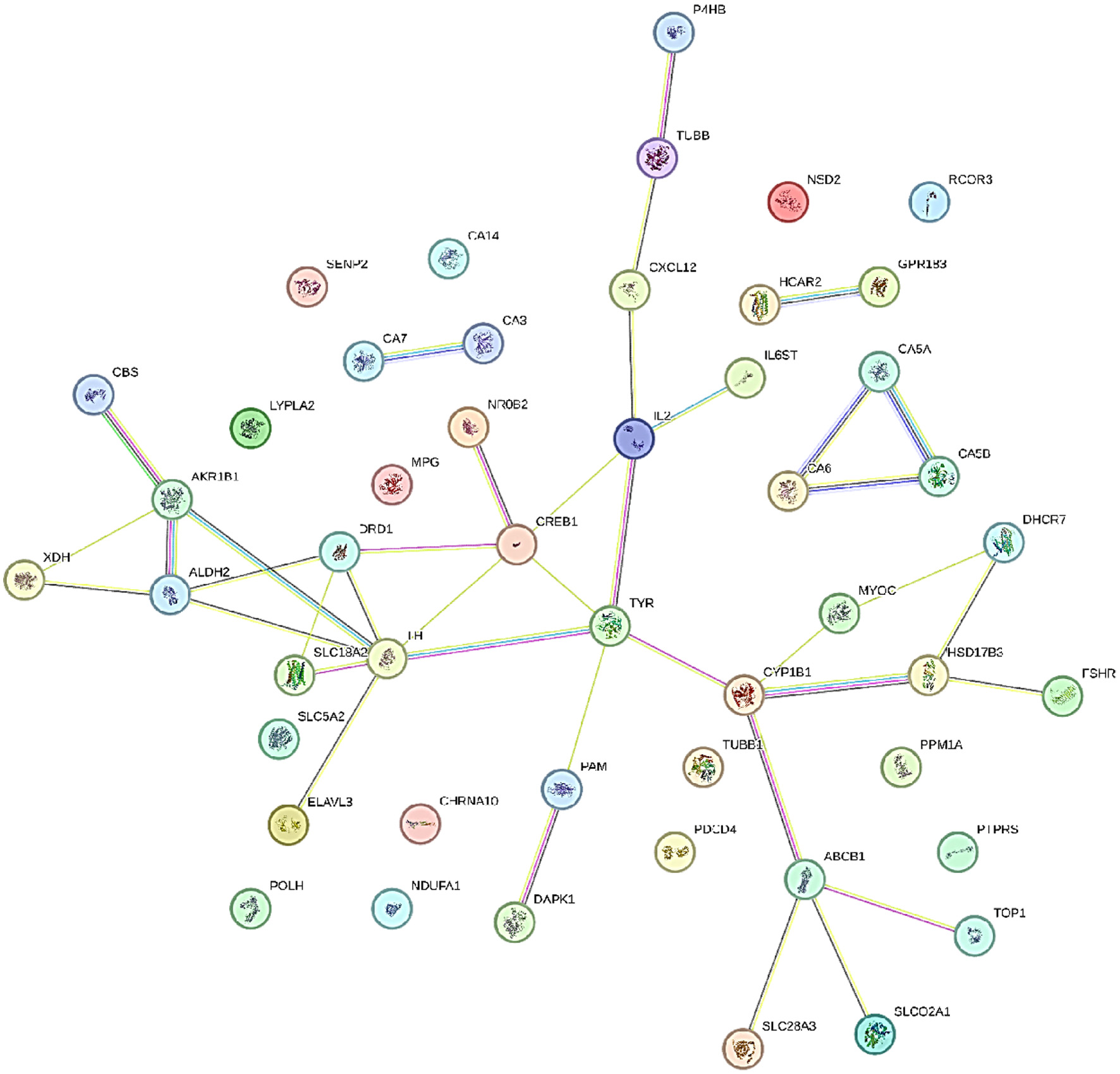
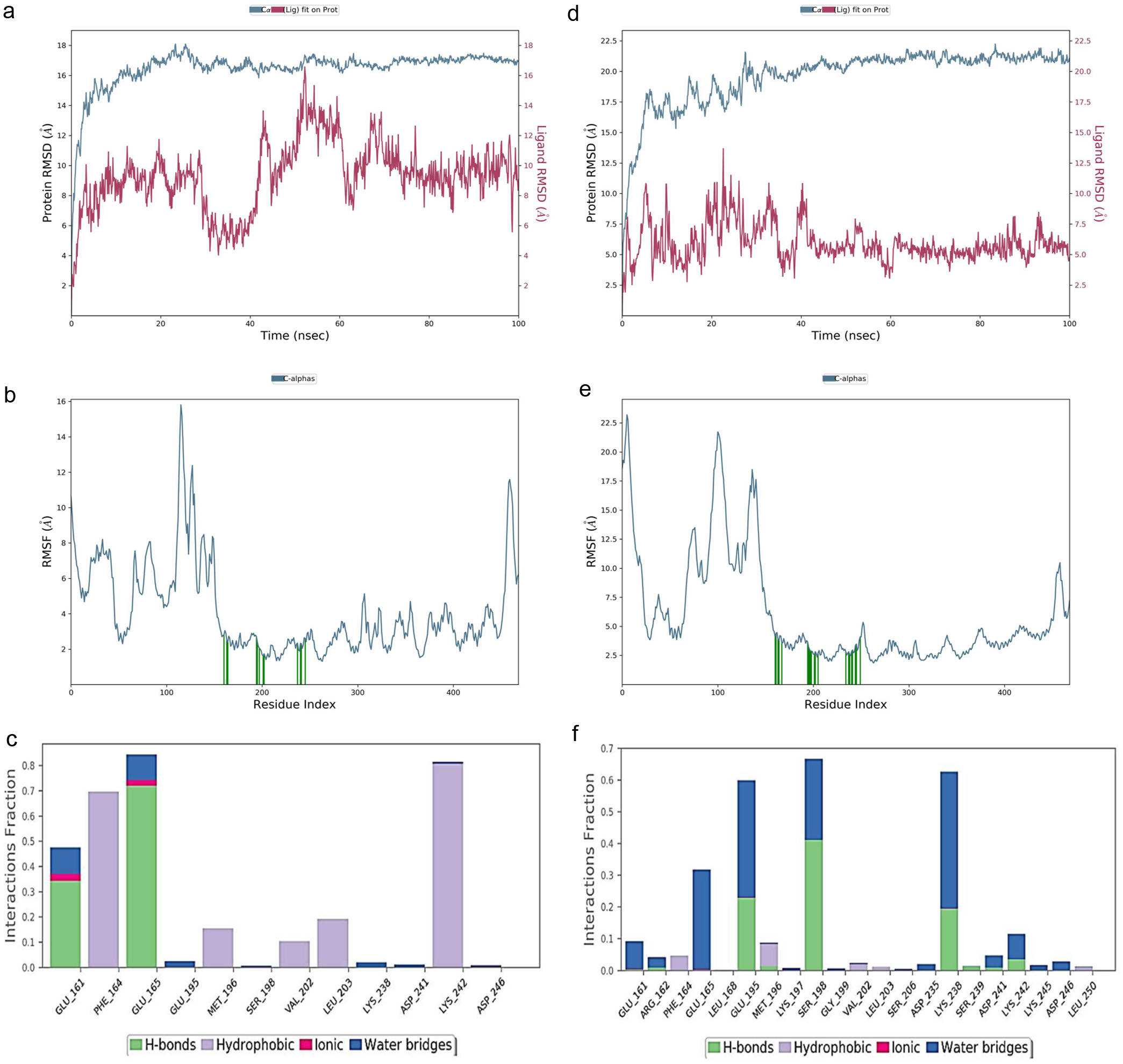
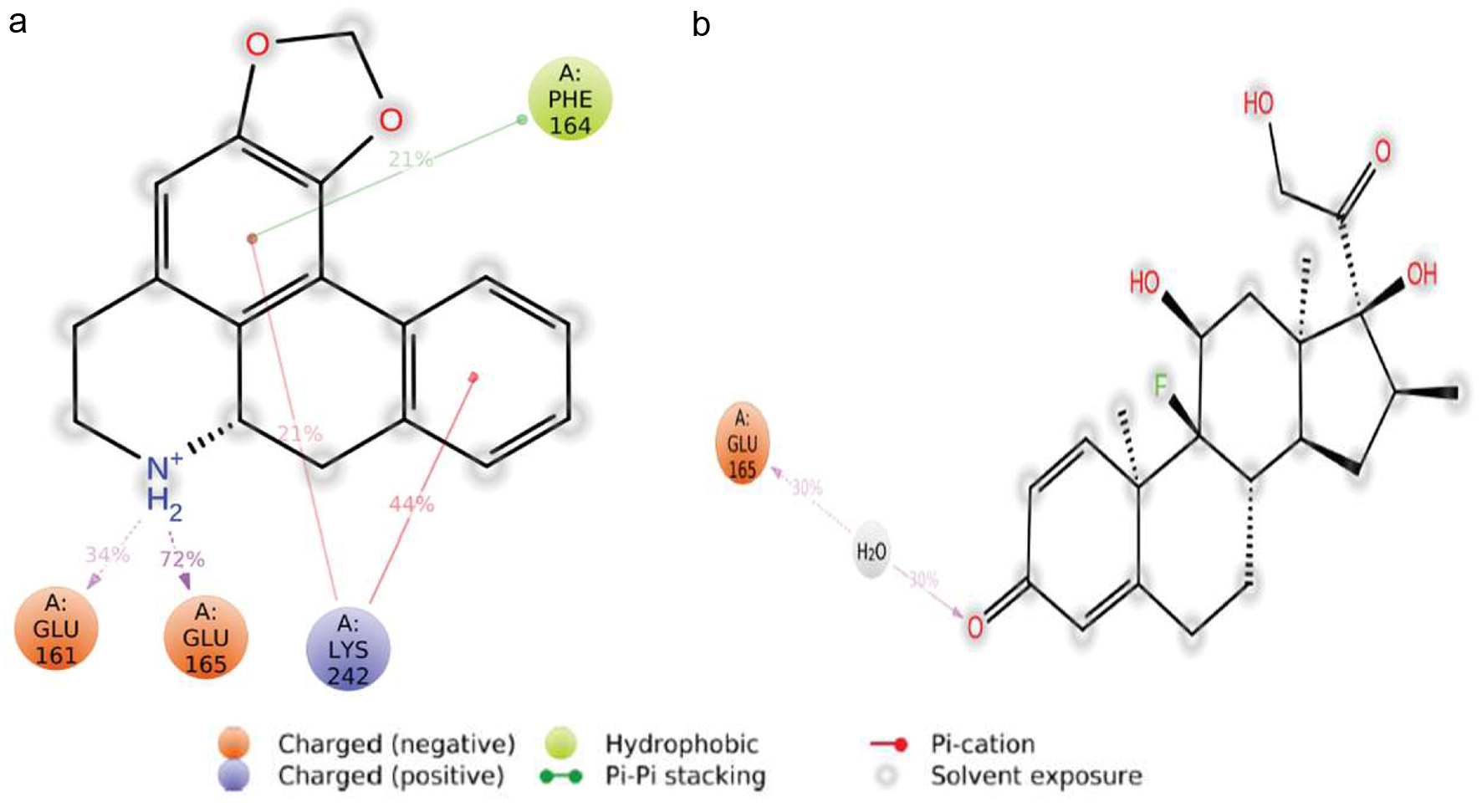
Figure 1. Protein-protein interaction of A. muricata molecular targets.
| Journal of Food Bioactives, ISSN 2637-8752 print, 2637-8779 online |
| Journal website www.isnff-jfb.com |
Original Research
Volume 25, March 2024, pages 81-94
In silico ADME and molecular simulation studies of pharmacological activities of phytoconstituents of Annona muricata (L.) Fruit
Figures

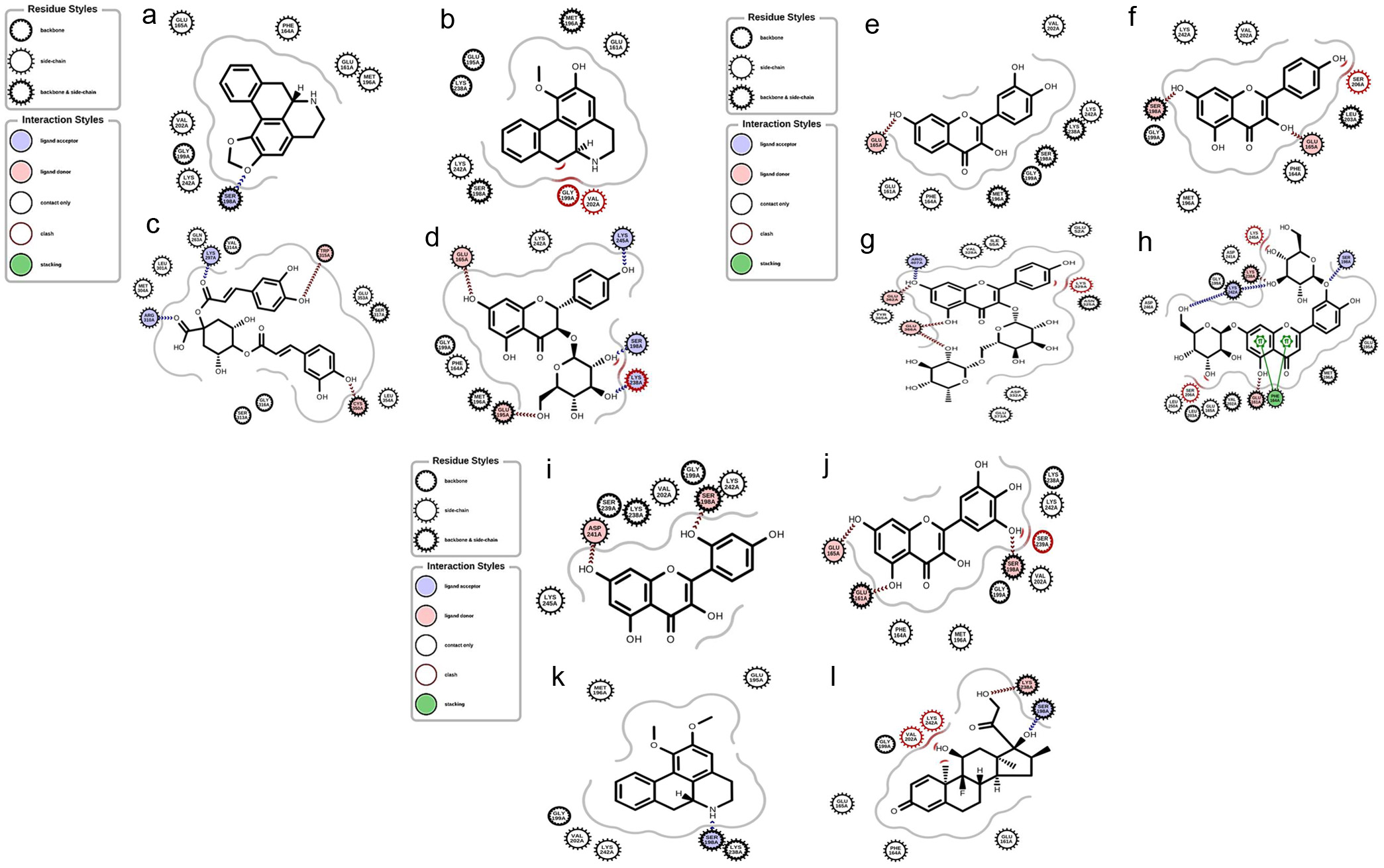
Tables
| SN | Ligands | Predicted ADME Parameter from SWISSADME | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MW | MR | TPSA (Å2) | Log P | ESOL Log S | ESOL Class | GIA | BBB permeant | P-gp | CYPs inhibitor | Lip | BS | SA | ||
| Physicochemical properties: Molecular weight (MW), Molar Refractivity (MR), Total polar surface area (TPSA). Lipophilicity: Consensus Log P. Water Solubility: ESOL Log S, ESOL Class. Pharmacokinetics: Gastrointestinal absorption (GIA), Blood-brain barrier (BBB), P-glycoprotein (P-gp) substrate, Inhibition of Cytochrome P450 (CYPs) type CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4. Druglikeness: Lipinski (Lip), Bioavailability Score (BS), Medicinal Chemistry: Synthetic accessibility (SA). | ||||||||||||||
| 1 | Annonacin | 596.88 | 172.67 | 116.45 | 6.92 | −7.13 | Poorly soluble | Low | No | No | CYP3A4 | 1 | 0.55 | 7.36 |
| 2 | Annonacin-10-one | 594.86 | 171.71 | 113.29 | 6.83 | −6.8 | Poorly soluble | Low | No | Yes | CYP3A4 | 1 | 0.55 | 7.18 |
| 3 | Annonaine | 265.31 | 80.13 | 30.49 | 2.88 | −3.71 | Soluble | High | Yes | Yes | CYP1A2, CYP2D6, CYP3A4 | 0 | 0.55 | 3.36 |
| 4 | Cis-Annoreticuin | 596.88 | 172.67 | 116.45 | 6.94 | −7.13 | Poorly soluble | Low | No | No | CYP3A4 | 1 | 0.55 | 7.36 |
| 5 | Asimilobine | 267.32 | 82.59 | 41.49 | 2.65 | −3.54 | Soluble | High | Yes | Yes | CYP1A2, CYP2D6, CYP3A4 | 0 | 0.55 | 3.24 |
| 6 | Cinnamic acid | 148.16 | 43.11 | 37.3 | 1.79 | −2.37 | Soluble | High | Yes | No | NONE | 0 | 0.85 | 1.67 |
| 7 | Corossolone | 578.86 | 170.55 | 93.06 | 7.75 | −7.66 | Poorly soluble | Low | No | Yes | NONE | 2 | 0.17 | 6.9 |
| 8 | Coumaric acid | 164.16 | 45.13 | 57.53 | 1.26 | −2.02 | Soluble | High | Yes | No | NONE | 0 | 0.85 | 1.61 |
| 9 | Dicaffeoylquinic acid | 516.45 | 126.9 | 211.28 | 0.91 | −3.65 | Soluble | Low | No | Yes | NONE | 3 | 0.11 | 4.83 |
| 10 | Dihydrokaempferol-hexoside | 450.39 | 105.12 | 186.37 | −0.5 | −2.8 | Soluble | Low | No | Yes | NONE | 2 | 0.17 | 5.24 |
| 11 | Epoxymurin-A | 530.86 | 167.55 | 38.83 | 10.35 | −10.03 | Insoluble | Low | No | Yes | NONE | 2 | 0.17 | 6.47 |
| 12 | Epoxymurin-B | 530.86 | 167.55 | 38.83 | 10.37 | −10.03 | Insoluble | Low | No | Yes | NONE | 2 | 0.17 | 6.47 |
| 13 | Epomusenin-A | 558.92 | 177.17 | 38.83 | 11.09 | −10.75 | Insoluble | Low | No | Yes | NONE | 2 | 0.17 | 6.73 |
| 14 | Epomusenin-B | 558.92 | 177.17 | 38.83 | 11.08 | −10.75 | Insoluble | Low | No | Yes | NONE | 2 | 0.17 | 6.73 |
| 15 | Fisetin | 286.24 | 76.01 | 111.13 | 1.55 | −3.35 | Soluble | High | No | No | CYP1A2, CYP2D6, CYP3A4 | 0 | 0.55 | 3.16 |
| 16 | Kaempferol | 286.24 | 76.01 | 111.13 | 1.58 | −3.31 | Soluble | High | No | No | CYP1A2, CYP2D6, CYP3A4 | 0 | 0.55 | 3.14 |
| 17 | Kaempferol 3-O-rutinoside | 594.52 | 139.36 | 249.2 | −0.73 | −3.42 | Soluble | Low | No | Yes | NONE | 3 | 0.17 | 6.48 |
| 18 | Luteolin 3,7-di-O-glucoside | 610.52 | 140.26 | 269.43 | −1.49 | −3.22 | Soluble | Low | No | Yes | NONE | 3 | 0.17 | 6.39 |
| 19 | Montecristin | 574.92 | 180.05 | 66.76 | 9.92 | −9.64 | Poorly soluble | Low | No | Yes | NONE | 2 | 0.17 | 6.82 |
| 20 | Morin | 302.24 | 78.03 | 131.36 | 1.2 | −3.16 | Soluble | High | No | No | CYP1A2, CYP2D6, CYP3A4 | 0 | 0.55 | 3.25 |
| 21 | Muricatetrocin B | 470.64 | 129.41 | 116.45 | 3.8 | −4.1 | Moderately soluble | High | No | No | CYP3A4 | 0 | 0.55 | 6.21 |
| 22 | Muricatocin A | 612.88 | 173.84 | 136.68 | 6.17 | −6.61 | Poorly soluble | Low | No | No | CYP3A4 | 1 | 0.55 | 7.5 |
| 23 | Myricetin | 318.24 | 80.06 | 151.59 | 0.79 | −3.01 | Soluble | Low | No | No | CYP1A2, CYP3A4 | 1 | 0.55 | 3.27 |
| 24 | N-methylcoclaurine | 299.36 | 90.52 | 52.93 | 2.59 | −3.82 | Soluble | High | Yes | Yes | CYP2D6 | 0 | 0.55 | 2.92 |
| 25 | Nornuciferine | 281.35 | 87.06 | 30.49 | 3.01 | −3.74 | Soluble | High | Yes | Yes | CYP1A2, CYP2D6, CYP3A4 | 0 | 0.55 | 3.35 |
| 26 | Reticuline | 329.39 | 97.01 | 62.16 | 2.6 | −3.88 | Soluble | High | Yes | Yes | CYP2D6 | 0 | 0.55 | 3.07 |
| 27 | Sabadelin | 530.86 | 167.55 | 38.83 | 10.35 | −10.03 | Insoluble | Low | No | Yes | NONE | 2 | 0.17 | 6.47 |
| 28 | Xylomatenin | 622.92 | 181.81 | 116.45 | 7.47 | −7.43 | Poorly soluble | Low | No | No | CYP3A4 | 1 | 0.55 | 7.58 |
| SN | Compound | Target gene code | Target description | p-value | MTC |
|---|---|---|---|---|---|
| 1 | Annonacin | PDCD4 | Programmed cell death protein 4 | 4.236e-74 | 0.36 |
| PTGER2 | Prostaglandin E2 receptor EP2 subtype | 1.586e-06 | 0.32 | ||
| 2 | Annonacin-10-one | PDCD4 | Programmed cell death protein 4 | 6.804e-70 | 0.33 |
| IL6ST | Interleukin-6 receptor subunit beta | 4.887e-23 | 0.30 | ||
| PPM1A | Protein phosphatase 1A | 7.318e-23 | 0.28 | ||
| SLCO2A1 | Solute carrier organic anion transporter family member 2A1 | 8.104e-18 | 0.28 | ||
| 3 | Annonaine | TAS1R1 | Taste receptor type 1 member 1 | 3.462e-06 | 0.29 |
| 4 | Cis-Annoreticuin | PDCD4 | Programmed cell death protein 4 | 4.236e-74 | 0.36 |
| 5 | Asimilobine | TH | Tyrosine 3-monooxygenase | 1.001e-39 | 0.32 |
| DRD1 | D(1A) dopamine receptor | 1.11e-16 | 0.39 | ||
| MMP26 | Matrix metalloproteinase-26 | 1.384e-07 | 0.32 | ||
| 6 | Cinnamic acid | HCAR2 | Hydroxycarboxylic acid receptor 2 | 8.423e-10 | 1.00 |
| NR0B2 | Nuclear receptor subfamily 0 group B member 2 | 9.432e-46 | 0.31 | ||
| GPR183 | G-protein coupled receptor 183 | 2.941e-42 | 0.39 | ||
| SENP2 | Sentrin-specific protease 2 | 3.568e-41 | 0.33 | ||
| CYP1B1 | Cytochrome P450 1B1 | 6.792e-40 | 0.48 | ||
| RCOR3 | REST corepressor 3 | 1.157e-39 | 0.31 | ||
| PAM | Peptidyl-glycine alpha-amidating monooxygenase | 1.372e-38 | 0.52 | ||
| CHRNA10 | Neuronal acetylcholine receptor subunit alpha-10 | 1.097e-35 | 0.28 | ||
| 7 | Corossolone | PDCD4 | Programmed cell death protein 4 | 3.7e-33 | 0.30 |
| PPM1A | Protein phosphatase 1A | 2.063e-23 | 0.29 | ||
| 8 | Coumaric acid | CA3 | Carbonic anhydrase 3 | 1.966e-29 | 1.00 |
| CA6 | Carbonic anhydrase 6 | 4.201e-29 | 1.00 | ||
| CA5B | Carbonic anhydrase 5B, mitochondrial | 5.523e-27 | 1.00 | ||
| CA5A | Carbonic anhydrase 5A, mitochondrial | 2.466e-26 | 1.00 | ||
| CA14 | Carbonic anhydrase 14 | 7.845e-21 | 1.00 | ||
| CA7 | Carbonic anhydrase 7 | 9.975e-20 | 1.00 | ||
| AKR1B1 | Aldo-keto reductase family 1 member B1 | 5.551e-16 | 1.00 | ||
| 9 | Dicaffeoylquinic acid | NSD2 | Histone-lysine N-methyltransferase NSD2 | 0.0008982 | 1.00 |
| CXCL12 | Stromal cell-derived factor 1 | 1.437e-76 | 0.38 | ||
| NR0B2 | Nuclear receptor subfamily 0 group B member 2 | 3.814e-47 | 0.33 | ||
| MYOC | Myocilin | 1.277e-31 | 0.39 | ||
| 10 | Dihydrokaempferol-hexoside | TOP1 | DNA topoisomerase 1 | 6.893e-61 | 0.36 |
| SLC5A2 | Sodium/glucose cotransporter 2 | 8.932e-58 | 0.43 | ||
| SLC28A3 | Solute carrier family 28 member 3 | 9.656e-53 | 0.42 | ||
| TYR | Tyrosinase | 5.641e-36 | 0.42 | ||
| IL2 | Interleukin-2 | 5.276e-30 | 0.40 | ||
| CBS | Cystathionine beta-synthase | 2.038e-27 | 0.48 | ||
| 11 | Epoxymurin-A | PDCD4 | Programmed cell death protein 4 | 1.016e-45 | 0.36 |
| LYPLA2 | Acyl-protein thioesterase 2 | 1.4e-22 | 0.29 | ||
| 12 | Epoxymurin-B | PDCD4 | Programmed cell death protein 4 | 1.016e-45 | 0.36 |
| LYPLA2 | Acyl-protein thioesterase 2 | 1.4e-22 | 0.29 | ||
| 13 | Epomusenin-A | PDCD4 | Programmed cell death protein 4 | 1.016e-45 | 0.36 |
| LYPLA2 | Acyl-protein thioesterase 2 | 1.4e-22 | 0.29 | ||
| 14 | Epomusenin-B | PDCD4 | Programmed cell death protein 4 | 1.016e-45 | 0.36 |
| LYPLA2 | Acyl-protein thioesterase 2 | 1.4e-22 | 0.29 | ||
| 15 | Fisetin | ELAVL3 | ELAV-like protein 3 | 2.048e-65 | 0.66 |
| HSD17B3 | Testosterone 17-beta-dehydrogenase 3 | 1.047e-52 | 0.43 | ||
| ALDH2 | Aldehyde dehydrogenase, mitochondrial | 4.45e-50 | 0.41 | ||
| CYP1B1 | Cytochrome P450 1B1 | 6.994e-39 | 0.66 | ||
| 16 | Kaempferol | ELAVL3 | ELAV-like protein 3 | 1.407e-81 | 0.78 |
| PTPRS | Receptor-type tyrosine-protein phosphatase S | 5.492e-59 | 0.75 | ||
| CBS | Cystathionine beta-synthase | 9.017e-51 | 0.45 | ||
| CREB1 | Cyclic AMP-responsive element-binding protein 1 | 5.513e-45 | 0.30 | ||
| P4HB | Protein disulfide-isomerase | 6.101e-41 | 0.52 | ||
| 17 | Kaempferol 3-O-rutinoside | P4HB | Protein disulfide-isomerase | 3.535e-71 | 1.00 |
| SLC28A3 | Solute carrier family 28 member 3 | 1.353e-43 | 0.35 | ||
| ELAVL3 | ELAV-like protein 3 | 2.773e-43 | 0.39 | ||
| IL2 | Interleukin-2 | 1.262e-31 | 0.42 | ||
| TYR | Tyrosinase | 9.763e-30 | 0.34 | ||
| XDH | Xanthine dehydrogenase/oxidase | 1.157e-25 | 0.65 | ||
| 18 | Luteolin 3,7-di-O-glucoside | CREB1 | Cyclic AMP-responsive element-binding protein 1 | 5.266e-69 | 0.35 |
| SLC5A2 | Sodium/glucose cotransporter 2 | 3.962e-60 | 0.41 | ||
| SLC28A3 | Solute carrier family 28 member 3 | 3.077e-50 | 0.40 | ||
| IL2 | Interleukin-2 | 5.836e-40 | 0.89 | ||
| 19 | Montecristin | PDCD4 | Programmed cell death protein 4 | 1.128e-77 | 0.41 |
| SLCO2A1 | Solute carrier organic anion transporter family member 2A1 | 2.172e-38 | 0.32 | ||
| POLH | DNA polymerase eta | 3.331e-16 | 0.31 | ||
| 20 | Morin | PTPRS | Receptor-type tyrosine-protein phosphatase S | 6.131e-56 | 1.00 |
| MPG | DNA-3-methyladenine glycosylase | 1.089e-50 | 1.00 | ||
| DAPK1 | Death-associated protein kinase 1 | 1.344e-21 | 1.00 | ||
| ELAVL3 | ELAV-like protein 3 | 8.402e-82 | 0.68 | ||
| P4HB | Protein disulfide-isomerase | 3.742e-36 | 0.44 | ||
| 21 | Muricatetrocin B | PDCD4 | Programmed cell death protein 4 | 2.688e-59 | 0.33 |
| 22 | Muricatocin A | PDCD4 | Programmed cell death protein 4 | 1.009e-81 | 0.40 |
| 23 | Myricetin | ELAVL3 | ELAV-like protein 3 | 2.196e-101 | 1.00 |
| XDH | Xanthine dehydrogenase/oxidase | 5.844e-36 | 1.00 | ||
| 24 | N-methylcoclaurine | DRD1 | D(1A) dopamine receptor | 1.982e-95 | 0.58 |
| DHCR7 | 7-dehydrocholesterol reductase | 2.187e-43 | 0.50 | ||
| TUBB1 | Tubulin beta-1 chain | 5.991e-26 | 0.35 | ||
| ABCB1 | ATP-dependent translocase ABCB1 | 9.037e-26 | 0.45 | ||
| SLC18A2 | Synaptic vesicular amine transporter | 4.29e-24 | 0.35 | ||
| 25 | Nornuciferine | TUBB | Tubulin beta chain | 4.386e-25 | 0.31 |
| 26 | Reticuline | DRD1 | D(1A) dopamine receptor | 9.256e-98 | 0.56 |
| DHCR7 | 7-dehydrocholesterol reductase | 3.993e-44 | 0.51 | ||
| TUBB1 | Tubulin beta-1 chain | 1.432e-43 | 0.40 | ||
| FSHR | Follicle-stimulating hormone receptor | 1.151e-29 | 0.30 | ||
| NDUFA1 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1 | 5.531e-17 | 0.32 | ||
| 27 | Sabadelin | PDCD4 | Programmed cell death protein 4 | 1.016e-45 | 0.36 |
| LYPLA2 | Acyl-protein thioesterase 2 | 1.4e-22 | 0.29 | ||
| 28 | Xylomatenin | PDCD4 | Programmed cell death protein 4 | 9.378e-70 | 0.41 |
| SLCO2A1 | Solute carrier organic anion transporter family member 2A1 | 1.494e-36 | 0.31 |
| S.N | Phytochemicals | PubChem CID | PDCD4 (AlphaFold ID: AF-Q53EL6) Binding Affinity ΔG (kcal.mol−1) |
|---|---|---|---|
| Docking parameter: PDCD4 [spacing: 0.525, box size: 126 × 126 × 126, center: −9.248 × 0.161 × 1.314]. | |||
| 1 | Annonacin | 354398 | −4.741 |
| 2 | Annonacin-10-one | 180161 | −4.591 |
| 3 | Annonaine | 160597 | −7.294 |
| 4 | Cis-Annoreticuin | 72778911 | −4.869 |
| 5 | Asimilobine | 160875 | −6.409 |
| 6 | Cinnamic acid | 444539 | −3.96 |
| 7 | Corossolone | 4366126 | −4.615 |
| 8 | Coumaric acid | 637542 | −5.183 |
| 9 | Dicaffeoylquinic acid | 12358846 | −6.775 |
| 10 | Dihydrokaempferol-hexoside | 10478918 | −7.012 |
| 11 | Epoxymurin-A | 5281161 | −4.416 |
| 12 | Epoxymurin-B | 131752983 | −3.789 |
| 13 | Epomusenin-A | 10507050 | −3.842 |
| 14 | Epomusenin-B | 10698082 | −3.614 |
| 15 | Fisetin | 5281614 | −6.979 |
| 16 | Kaempferol | 5280863 | −6.749 |
| 17 | Kaempferol 3-O-rutinoside | 5318767 | −6.629 |
| 18 | Luteolin 3,7-di-O-glucoside | 5490298 | −7.65 |
| 19 | Montecristin | 102083640 | −3.861 |
| 20 | Morin | 5281670 | −6.884 |
| 21 | Muricatetrocin B | 393472 | −5.362 |
| 22 | Muricatocin A | 133072 | −5.191 |
| 23 | Myricetin | 5281672 | −6.538 |
| 24 | N-methylcoclaurine | 440595 | −5.946 |
| 25 | Nornuciferine | 41169 | −6.226 |
| 26 | Reticuline | 439653 | −5.907 |
| 27 | Sabadelin | 101006011 | −3.426 |
| 28 | Xylomatenin | 10484035 | −4.5 |
| STD | Dexamethasone | 5743 | −6.682 |
| Complex | Simulation Time (ns) | MMGBSA ΔGbind (kcal.mol−1) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Coulomb | Covalent | Hbond | Lipo | Packing | Solv_GB | vdW | Gbind (Total) | ||
| Covalent: Covalent binding energy; Coulomb: Coulomb energy; Lipo: Lipophilic energy; Hbond: Hydrogen bonding energy; Packing: Pi-pi packing correction; Solv GB: Generalized Born electrostatic solvation energy; vdW: Van der Waals energy; Total: Total energy (Prime energy). | |||||||||
| Annonaine and PDCD4 | 0 | −64.428 | 2.791 | −0.60 | −14.323 | −0.541 | 69.403 | −28.149 | −35.851 |
| 100 | −52.471 | 1.676 | −0.797 | −15.184 | −1.519 | 52.998 | −23.720 | −39.019 | |
| Dexamethasone and PDCD4 | 0 | 0.913 | 0.092 | −0.420 | −7.605 | 0 | 11.380 | −32.850 | −28.489 |
| 100 | −4.693 | −0.012 | −0.027 | −8.555 | 0 | 15.696 | −30.694 | −28.284 | |